 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013592301.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 782aa MW: 85891 Da PI: 6.8148 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 61 | 1.8e-19 | 100 | 155 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ +++t++q++ Le++F+++ +p++++r +L+++l+L+ rqVk+WFqNrR+++k
XP_013592301.1 100 KKRYHRHTAKQIQDLESVFKECAHPDEKQRLDLSRRLNLDPRQVKFWFQNRRTQMK 155
688999***********************************************999 PP
| |||||||
| 2 | START | 184 | 7.9e-58 | 300 | 519 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEE CS
START 2 laeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv.....dsgealrasgvvdmvlallveellddkeqWdetla....kaetle 83
la+ a++elvk+a ep+Wv+ss e++n++e+ ++f++ + + +ea++++g+v+ ++ lve+l+d+ +W e+++ + +t+e
XP_013592301.1 300 LALGAMEELVKMARRHEPLWVRSSetgfEMLNKEEYDTSFSRVVGpkqdgFVSEASKETGNVIINSLALVETLMDSE-RWAEMFPsmisRTSTTE 393
67889***********************88888888888866544888999**************************.*******9999****** PP
EECTT......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE CS
START 84 vissg......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehvd 171
issg gal+lm aelq+lsplvp R + f+R+++q+ +g+w++vdvS+ds ++ + sss+ R lpSg+l+++++ng+skvtw+eh++
XP_013592301.1 394 IISSGmggtrnGALHLMHAELQLLSPLVPvRQVSFLRFCKQHAEGVWAVVDVSIDSIREGS-SSSCRR---LPSGCLVQDMANGYSKVTWIEHTE 484
************************************************************9.777766...************************ PP
--SSXXHHHHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 172 lkgrlphwllrslvksglaegaktwvatlqrqcek 206
++++ +h l+r+l++ gla+ga++w+a+lqrqce+
XP_013592301.1 485 YDENRIHRLYRPLLSCGLAFGAQRWMAALQRQCEC 519
*********************************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-21 | 86 | 157 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 2.76E-19 | 88 | 157 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.216 | 97 | 157 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.5E-17 | 98 | 161 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.00E-16 | 100 | 157 | No hit | No description |
| Pfam | PF00046 | 4.5E-17 | 100 | 155 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 132 | 155 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 39.624 | 290 | 522 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 6.04E-30 | 293 | 519 | No hit | No description |
| CDD | cd08875 | 1.08E-111 | 294 | 518 | No hit | No description |
| SMART | SM00234 | 1.0E-44 | 299 | 519 | IPR002913 | START domain |
| Pfam | PF01852 | 2.1E-49 | 301 | 519 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 1.26E-18 | 549 | 774 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048497 | Biological Process | maintenance of floral organ identity | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 782 aa Download sequence Send to blast |
MNFNGYLGDG SSGDRISDVP YSDHVSFPAA ATMPHSRPFS SNGLSLGLQT NGVNGETFET 60 NVTRNKSRGG EDAESRSESD NAEAASGDDL ETGDKPPRKK KRYHRHTAKQ IQDLESVFKE 120 CAHPDEKQRL DLSRRLNLDP RQVKFWFQNR RTQMKTQIER HENALLRQEN DKLRAENMSV 180 REAMRNPMCS NCGGPAVPGE VSMEEQHLRI ENSRLKDELD RVCALTGKFF GRSPSGSHHV 240 PDSSLLLGVG VGSGGGFSLS SPSLPQASPR FEISNDTGLA TVNRQPPVSD FDQRSRYLDL 300 ALGAMEELVK MARRHEPLWV RSSETGFEML NKEEYDTSFS RVVGPKQDGF VSEASKETGN 360 VIINSLALVE TLMDSERWAE MFPSMISRTS TTEIISSGMG GTRNGALHLM HAELQLLSPL 420 VPVRQVSFLR FCKQHAEGVW AVVDVSIDSI REGSSSSCRR LPSGCLVQDM ANGYSKVTWI 480 EHTEYDENRI HRLYRPLLSC GLAFGAQRWM AALQRQCECL TILMSSTVSP SRSPTPISCN 540 GRKSMLKLAR RMTDNFCGGV CASSLQKWSK LNVGNVDEDV RIMTRKSVND PGEPPGIVLN 600 AATSVWMPVS PKRLFDFLGN ERLRSEWDIL SNGGPMQEMA HIAKGHDHSN SISLLRATAI 660 NANQSSMLIL QETSIDAAGA VVVYAPVDIP AMQTVMNGGD SAYVALLPSG FAILPNAPQR 720 LEERNGNGGC MAEGGSLLTV AFQILVNSLP TAKLTVESVE TVNNLISCTI QKIKAALHCD 780 SN |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in trichomes forming at the base of young leaves, in endodermal cell lines around emergent lateral roots and in the epidermal layer of the stamen filament. {ECO:0000269|PubMed:16778018}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00418 | DAP | Transfer from AT3G61150 | Download |
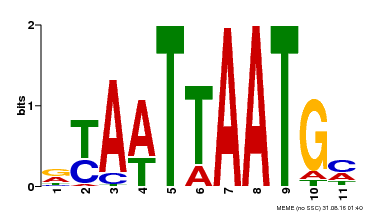
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013592301.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ182490 | 0.0 | DQ182490.1 Brassica napus baby boom interacting protein 2 mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013592301.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HDG1 | ||||
| Swissprot | Q9M2E8 | 0.0 | HDG1_ARATH; Homeobox-leucine zipper protein HDG1 | ||||
| TrEMBL | A0A0D3CV31 | 0.0 | A0A0D3CV31_BRAOL; Uncharacterized protein | ||||
| STRING | Bo6g077480.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1128 | 27 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G61150.1 | 0.0 | homeodomain GLABROUS 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



