 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013593880.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 219aa MW: 23219.8 Da PI: 9.2568 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 119.1 | 1.7e-37 | 24 | 83 | 3 | 62 |
zf-Dof 3 ekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
++l+cprC+stntkfCyynny+ sqPr+fCkaCrryWt+GG+lr++PvGg +rk+ k+s
XP_013593880.1 24 PDQLPCPRCESTNTKFCYYNNYNFSQPRHFCKACRRYWTHGGTLRDIPVGGVSRKSSKRS 83
56899**************************************************98875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 2.0E-31 | 2 | 75 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 7.0E-33 | 26 | 81 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.584 | 27 | 81 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 29 | 65 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 219 aa Download sequence Send to blast |
MPSDVNESRR LTKIPHGGAA ADQPDQLPCP RCESTNTKFC YYNNYNFSQP RHFCKACRRY 60 WTHGGTLRDI PVGGVSRKSS KRSRTCSSVP VSTAVGSREF PLQATPVLFP PSSNDGGVTA 120 AKGTASSAFG GFTSLLSYNA AASRNGPGGR FNGLEGFGLG LGQGSYFDNV RFGQGITVWP 180 FSTGATDATS AVTSHVGPIP ATWQFEGPES KVGFVSGDY |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00593 | DAP | Transfer from AT5G66940 | Download |
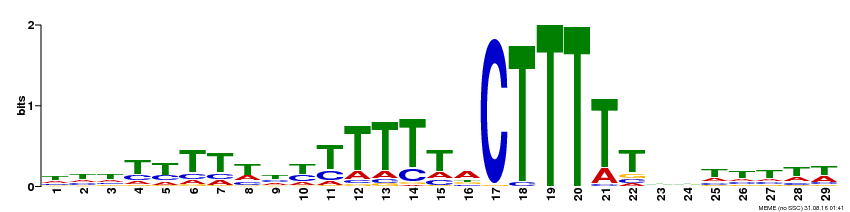
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013593880.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013593880.1 | 1e-161 | PREDICTED: dof zinc finger protein DOF5.8 | ||||
| Refseq | XP_013687059.1 | 1e-161 | dof zinc finger protein DOF5.8 | ||||
| Swissprot | Q9FGD6 | 1e-115 | DOF58_ARATH; Dof zinc finger protein DOF5.8 | ||||
| TrEMBL | A0A078FEI8 | 1e-159 | A0A078FEI8_BRANA; BnaC07g16430D protein | ||||
| TrEMBL | A0A0D3D8A9 | 1e-159 | A0A0D3D8A9_BRAOL; Uncharacterized protein | ||||
| TrEMBL | A0A3P6EJ53 | 1e-159 | A0A3P6EJ53_BRAOL; Uncharacterized protein | ||||
| STRING | Bo7g064260.1 | 1e-160 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM2286 | 28 | 74 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G66940.1 | 3e-99 | Dof family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



