 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013595944.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 368aa MW: 42619.7 Da PI: 5.3937 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 178.6 | 1.6e-55 | 10 | 139 | 1 | 129 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk..kvka.eekewyfFskrdkkyatgkrknratksgyWkatgkdkev 92
+ppGfrFhPtdeelv +yL+kkv+++k++l +vi+++d+y++ePwdL++ ++ e++ewyfFs++dkky+tg+r+nrat +g+Wkatg+dk+v
XP_013595944.1 10 VPPGFRFHPTDEELVGYYLRKKVASQKIDL-DVIRDIDLYRIEPWDLQErcRIGYeERNEWYFFSHKDKKYPTGTRTNRATMAGFWKATGRDKAV 103
69****************************.9***************953433332556************************************ PP
NAM 93 lskkgelvglkktLvfykgrapkgektdWvmheyrle 129
++ k++l+g++ktLvfykgrap+g+ktdW+mheyrle
XP_013595944.1 104 YE-KSKLMGMRKTLVFYKGRAPNGQKTDWIMHEYRLE 139
**.999*****************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 1.57E-60 | 7 | 159 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 59.441 | 10 | 159 | IPR003441 | NAC domain |
| Pfam | PF02365 | 9.6E-29 | 11 | 138 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048759 | Biological Process | xylem vessel member cell differentiation | ||||
| GO:1901348 | Biological Process | positive regulation of secondary cell wall biogenesis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 368 aa Download sequence Send to blast |
MESVDQSCSV PPGFRFHPTD EELVGYYLRK KVASQKIDLD VIRDIDLYRI EPWDLQERCR 60 IGYEERNEWY FFSHKDKKYP TGTRTNRATM AGFWKATGRD KAVYEKSKLM GMRKTLVFYK 120 GRAPNGQKTD WIMHEYRLES DENAPPQEEG WVVCRAFKKK PTTGQARNTE TWRSGYFYDQ 180 LPSGVGSSME PLNYVSKQKQ NIFAQDFMFK QELGGSDIGL NFIDCDQFIQ LPQLESPSFP 240 LMKRPVSSTS ITSLEKNHNK YKKPLTEDDE NFDELIKSKD KKKKASVMTT DWRALDKFVA 300 SQLMSQEDGV PDFGAHQEDD TNKTGQCNNE ESNNNGIEMP SSSLLSDREE ENRFISGFLC 360 TNLDYDLI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 4e-51 | 4 | 160 | 9 | 169 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Up-regulated during xylem vessel element formation. Expressed preferentially in procambial cells adjacent to root meristem. {ECO:0000269|PubMed:16103214, ECO:0000269|PubMed:25148240}. | |||||
| Uniprot | TISSUE SPECIFICITY: Detected in root protoxylem and metaxylem poles and in vessels of protoxylems, outermost metaxylems, inner metaxylems, shoots and hypocotyls (PubMed:18445131). Expressed in roots, hypocotyls, cotyledons and leaves. Present in developing xylems (PubMed:16103214, PubMed:16581911). Specifically expressed in vessels but not in interfascicular fibers in stems (PubMed:25148240). {ECO:0000269|PubMed:16103214, ECO:0000269|PubMed:16581911, ECO:0000269|PubMed:18445131, ECO:0000269|PubMed:25148240}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the secondary wall NAC binding element (SNBE), 5'-(T/A)NN(C/T)(T/C/G)TNNNNNNNA(A/C)GN(A/C/T)(A/T)-3', in the promoter of target genes (By similarity). Involved in xylem formation by promoting the expression of secondary wall-associated transcription factors and of genes involved in secondary wall biosynthesis and programmed cell death, genes driven by the secondary wall NAC binding element (SNBE). Triggers thickening of secondary walls (PubMed:25148240). {ECO:0000250|UniProtKB:Q9LVA1, ECO:0000269|PubMed:25148240}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00471 | DAP | Transfer from AT4G36160 | Download |
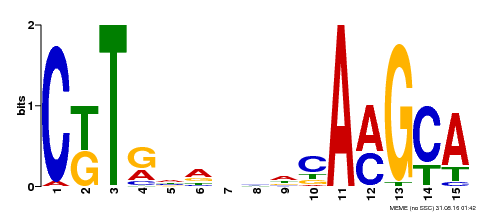
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013595944.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC189569 | 0.0 | AC189569.2 Brassica rapa subsp. pekinensis cultivar Inbred line 'Chiifu' clone KBrH009I12, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013595944.1 | 0.0 | PREDICTED: NAC domain-containing protein 76-like | ||||
| Refseq | XP_013595945.1 | 0.0 | PREDICTED: NAC domain-containing protein 76-like | ||||
| Swissprot | O65508 | 0.0 | NAC76_ARATH; NAC domain-containing protein 76 | ||||
| TrEMBL | A0A0D3DHY4 | 0.0 | A0A0D3DHY4_BRAOL; Uncharacterized protein | ||||
| TrEMBL | A0A3P6EY50 | 0.0 | A0A3P6EY50_BRAOL; Uncharacterized protein | ||||
| STRING | Bo7g117820.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3686 | 26 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36160.1 | 0.0 | NAC domain containing protein 76 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



