|
Plant Transcription
Factor Database
|
Transcription Factor Information
|
Basic
Information? help
Back to Top |
| TF ID |
XP_013602805.1 |
| Organism |
|
| Taxonomic ID |
|
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
| Family |
SRS |
| Protein Properties |
Length: 365aa MW: 37767.5 Da PI: 7.7531 |
| Description |
SRS family protein |
| Gene Model |
| Gene Model ID |
Type |
Source |
Coding Sequence |
| XP_013602805.1 | genome | NCBI | View CDS |
|
| Signature Domain? help Back to Top |
 |
| No. |
Domain |
Score |
E-value |
Start |
End |
HMM Start |
HMM End |
| 1 | DUF702 | 226.7 | 4.4e-70 | 137 | 286 | 4 | 152 |
DUF702 4 gtasCqdCGnqakkdCaheRCRtCCksrgfdCathvkstWvpaakrrerqqqlaaasskaaasa.aeaaskrkrelkskkqsalsstklssaesk 97
+++sCqdCGnqakkdCah RCRtCCksrgf+C thv+stWvpaakrrerqqql + + ++ + ++ +kr re+ +++s+l +t++++ ++
XP_013602805.1 137 TGVSCQDCGNQAKKDCAHVRCRTCCKSRGFQCPTHVRSTWVPAAKRRERQQQLGTVQPQTHLPRgDSGVPKRLRENLPATSSSLVCTRVPTYHNA 231
4679*************************************************999877766665788999************************ PP
DUF702 98 keletsslPeevsseavfrcvrvssvddgeeelaYqtavsigGhvfkGiLydqGl 152
+ le+ ++P+evss+avfrcvrvssv+dgeee+aYqtavsigGhvfkGiLyd+G+
XP_013602805.1 232 SGLEVGDFPAEVSSPAVFRCVRVSSVEDGEEEFAYQTAVSIGGHVFKGILYDNGP 286
******************************************************8 PP
|
| Sequence ? help Back to Top |
| Protein Sequence Length: 365 aa
Download sequence Send
to blast |
MAGFFSLDGG GGRGGGGNGQ EEHRTNTNPP PPVSESWLWY RNPNVNANAN APSSSNAAAL 60
GTLELWQNHN QQEIMFQHQQ HQQRLDLYSS AAGLGVGPSS HSQFNISGET SNAGAGGTAA 120
AALMMMRSGG SGGGGGTGVS CQDCGNQAKK DCAHVRCRTC CKSRGFQCPT HVRSTWVPAA 180
KRRERQQQLG TVQPQTHLPR GDSGVPKRLR ENLPATSSSL VCTRVPTYHN ASGLEVGDFP 240
AEVSSPAVFR CVRVSSVEDG EEEFAYQTAV SIGGHVFKGI LYDNGPGSIG GGSYNVGESS 300
SGGGGAQQMN LITAGSVTVA TASSSTPNAG GIGGSSAAYT DPAALYPTPI NTFMAGTQFF 360
PNPRS
|
| Expression --
Description ? help
Back to Top |
| Source |
Description |
| Uniprot | DEVELOPMENTAL STAGE: Expressed in the apical parts of the developing gynoecium. Detected throughout the youngest flower primordium. Later relocalizes towards the regions of the presumptive sepal anlagen and remains in sepal primordia until just after their emergence. Also observed on the abaxial side of the young floral meristem. Present in the newly arisen gynoecial primordium. Restricted to the apical parts of the carpels as the open-ended gynoecial cylinder elongates vertically. In the apical regions of the gynoecium, confined to a zone in the interphase between the style and the stigma and fades out later. Within the gynoecium, accumulates in ovule primordia, and, as the ovules developed, restricted to the epidermis of the developing funiculi, to the outer, but not the inner, integuments and to the tip of the nucellus. Also present in the cell layer of the septum that faces the ovary. In the embryo, detected in the cotyledon primordia during late globular to mid heart stage. In addition, transiently expressed in petal and stamen primordia and in the tapetum of the anthers. {ECO:0000269|PubMed:12361963}. |
| Uniprot | TISSUE SPECIFICITY: Expressed in flowers, seeds and seedlings. {ECO:0000269|PubMed:12361963}. |
| Functional Description ? help
Back to Top |
| Source |
Description |
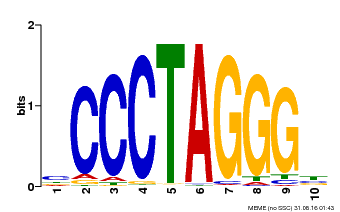
| UniProt | Transcription activator that binds DNA on 5'-ACTCTAC-3' and promotes auxin homeostasis-regulating gene expression (e.g. YUC genes), as well as genes affecting stamen development, cell expansion and timing of flowering. Synergistically with other SHI-related proteins, regulates gynoecium, stamen and leaf development in a dose-dependent manner, controlling apical-basal patterning. Promotes style and stigma formation, and influences vascular development during gynoecium development. May also have a role in the formation and/or maintenance of the shoot apical meristem (SAM). {ECO:0000269|PubMed:12361963, ECO:0000269|PubMed:16740145, ECO:0000269|PubMed:16740146, ECO:0000269|PubMed:18811619, ECO:0000269|PubMed:20154152, ECO:0000269|PubMed:22318676}. |
| Regulation -- Description ? help
Back to Top |
| Source |
Description |
| UniProt | INDUCTION: Regulated by ESR1 and ESR2. {ECO:0000269|PubMed:21976484}. |
| Annotation --
Nucleotide ? help
Back to Top |
| Source |
Hit ID |
E-value |
Description |
| GenBank | GU205262 | 0.0 | GU205262.1 Brassica rapa subsp. pekinensis stylish (STY1-1) mRNA, complete cds. |
| Best hit in Arabidopsis thaliana ? help
Back to Top |
| Hit ID |
E-value |
Description |
| AT3G51060.1 | 1e-149 | Lateral root primordium (LRP) protein-related |