 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013605597.1 | ||||||||
| Common Name | 23.t00048 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 586aa MW: 64243.5 Da PI: 4.9394 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 39.5 | 1e-12 | 415 | 460 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
+h e+Er+RR+++N++f Lr ++P+ K++Ka+ L A+ YI++L
XP_013605597.1 415 NHVEAERQRREKLNQRFYSLRAVVPNV-----SKMDKASLLGDAISYINEL 460
799***********************6.....5***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 1.3E-57 | 62 | 256 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 17.282 | 411 | 460 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.3E-18 | 414 | 481 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 4.20E-14 | 414 | 465 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 4.0E-19 | 415 | 480 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 3.8E-10 | 415 | 460 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 8.3E-17 | 417 | 466 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006952 | Biological Process | defense response | ||||
| GO:0009718 | Biological Process | anthocyanin-containing compound biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043425 | Molecular Function | bHLH transcription factor binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 586 aa Download sequence Send to blast |
MSSTNVQLTD HHLNQSTNGT NLWSTIEDNA SVMEPLIGSE HSSLWPQPPL TPPPPHVTED 60 TLQQRLQALI EGARESWTYA VFWQLSHDFA GEDISNTAAL LTWGDGYYKG EEERKSRKRK 120 PNPVSAAEQE HRKRVIRELN SLISGGGGTV SSSGGSSDEA GDEDVSDTEW FFLVSMTQSF 180 VNGSGLPGRA FSSSRTIWLS GSNALAGSSC ERARQGQVYG LETMVCIPTQ NGVVELGSLE 240 IIHQSSDLVE KVNSFFSFNG GGGGGESGSW EFNLNPDQGE NDSATWINEP IVTGIEPVLG 300 APATSNSDSQ TASKLCNGSS VEHPKQQQNP QISSSGFVEG DSNKKKRCLV SDKEEEMLSF 360 TSVLPLPTKS NDSNRSDLEA SVVKEAESGR IVAETEKKPR KRGRKPANGR EEPLNHVEAE 420 RQRREKLNQR FYSLRAVVPN VSKMDKASLL GDAISYINEL KAKLQKAEAD KEELQKQIDG 480 MSKEVGDGNV KSLVKDQKCL DQDSGVSIEV EIDVKIIGWD AMIRIQCAKK NHPGAKFMEA 540 LKELELEVNH ASLSVVNEFM IQQATVKMGN QFFTQDQLKA ALMERV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rqw_A | 7e-87 | 55 | 259 | 2 | 195 | Transcription factor MYC3 |
| 4rqw_B | 7e-87 | 55 | 259 | 2 | 195 | Transcription factor MYC3 |
| 4rru_A | 3e-86 | 17 | 252 | 1 | 227 | Transcription factor MYC3 |
| 4rs9_A | 7e-87 | 55 | 259 | 2 | 195 | Transcription factor MYC3 |
| 4ywc_A | 3e-86 | 17 | 252 | 1 | 227 | Transcription factor MYC3 |
| 4ywc_B | 3e-86 | 17 | 252 | 1 | 227 | Transcription factor MYC3 |
| 4yz6_A | 7e-87 | 55 | 259 | 2 | 195 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 396 | 404 | KKPRKRGRK |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed constitutively at low levels. Preferentially expressed in vascular tissues. {ECO:0000269|PubMed:12509522, ECO:0000269|PubMed:21335373}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
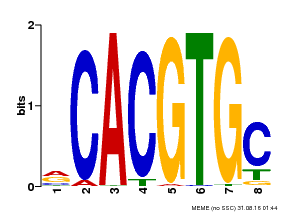
| UniProt | Transcription factor involved in jasmonic acid (JA) gene regulation. With MYC2 and MYC3, controls additively subsets of JA-dependent responses. Can form complexes with all known glucosinolate-related MYBs to regulate glucosinolate biosynthesis. Binds to the G-box (5'-CACGTG-3') of promoters. Activates multiple TIFY/JAZ promoters. {ECO:0000269|PubMed:21321051, ECO:0000269|PubMed:21335373, ECO:0000269|PubMed:23943862}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00086 | PBM | Transfer from AT4G17880 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013605597.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. Not induced by jasmonic acid. {ECO:0000269|PubMed:12679534, ECO:0000269|PubMed:21335373}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC183492 | 0.0 | AC183492.1 Brassica oleracea Contig G, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013605597.1 | 0.0 | PREDICTED: transcription factor MYC4 | ||||
| Swissprot | O49687 | 0.0 | MYC4_ARATH; Transcription factor MYC4 | ||||
| TrEMBL | A0A0D3A3X7 | 0.0 | A0A0D3A3X7_BRAOL; Uncharacterized protein | ||||
| TrEMBL | Q25BJ5 | 0.0 | Q25BJ5_BRAOL; Basic helix-loop-helix (BHLH) family transcription factor | ||||
| STRING | Bo1g017990.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1265 | 28 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G17880.1 | 0.0 | bHLH family protein | ||||



