 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013607098.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 330aa MW: 37289.5 Da PI: 8.679 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 99 | 6.8e-31 | 24 | 100 | 52 | 128 |
NAM 52 kaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlskkgelvglkktLvfykgrapkgektdWvmheyrl 128
+ +ekewyf+++rd+ky++g+r+nr+t+sgyWkatg d+ + s++ + +glkktLvfy+g+apkg++t+W+m+eyrl
XP_013607098.1 24 AIGEKEWYFYVPRDRKYRNGDRPNRVTTSGYWKATGADRMIRSESARPIGLKKTLVFYSGKAPKGTRTSWIMNEYRL 100
45789**********************************************************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51005 | 37.259 | 1 | 121 | IPR003441 | NAC domain |
| SuperFamily | SSF101941 | 5.89E-35 | 22 | 121 | IPR003441 | NAC domain |
| Pfam | PF02365 | 2.5E-13 | 28 | 100 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 330 aa Download sequence Send to blast |
MVSFKDYILL NVDYRCVLCA AMAAIGEKEW YFYVPRDRKY RNGDRPNRVT TSGYWKATGA 60 DRMIRSESAR PIGLKKTLVF YSGKAPKGTR TSWIMNEYRL PHHETEKYQK AEISLCRVYK 120 RPGVEDHLSL PRSLSTRHHN HTSSSSRLAL RQKQQHHSSS SNHSDNNLNN NNNLEKLSTE 180 YFGDGSSITT TNSNSDVTIA LANQNIYRPM PYGVSNTPMI STTNKEDDEN AIVDDLQRLV 240 NYQISDGGSN INHQYYQIAQ QFHNQQEVNA NALQLVAGAT TVALTPQTQA ALAMNMIPAG 300 TIPNSALWDL WNPLVPDGNR DHYTDSFQGI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ut4_A | 1e-35 | 23 | 121 | 66 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut4_B | 1e-35 | 23 | 121 | 66 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_A | 1e-35 | 23 | 121 | 66 | 165 | NO APICAL MERISTEM PROTEIN |
| 1ut7_B | 1e-35 | 23 | 121 | 66 | 165 | NO APICAL MERISTEM PROTEIN |
| 3swm_A | 1e-35 | 23 | 121 | 69 | 168 | NAC domain-containing protein 19 |
| 3swm_B | 1e-35 | 23 | 121 | 69 | 168 | NAC domain-containing protein 19 |
| 3swm_C | 1e-35 | 23 | 121 | 69 | 168 | NAC domain-containing protein 19 |
| 3swm_D | 1e-35 | 23 | 121 | 69 | 168 | NAC domain-containing protein 19 |
| 3swp_A | 1e-35 | 23 | 121 | 69 | 168 | NAC domain-containing protein 19 |
| 3swp_B | 1e-35 | 23 | 121 | 69 | 168 | NAC domain-containing protein 19 |
| 3swp_C | 1e-35 | 23 | 121 | 69 | 168 | NAC domain-containing protein 19 |
| 3swp_D | 1e-35 | 23 | 121 | 69 | 168 | NAC domain-containing protein 19 |
| 4dul_A | 1e-35 | 23 | 121 | 66 | 165 | NAC domain-containing protein 19 |
| 4dul_B | 1e-35 | 23 | 121 | 66 | 165 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in aerial organs in early stages of seedling development. {ECO:0000269|PubMed:17653269}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that acts as a floral repressor. Controls flowering time by negatively regulating CONSTANS (CO) expression in a GIGANTEA (GI)-independent manner. Regulates the plant cold response by positive regulation of the cold response genes COR15A and KIN1. May coordinate cold response and flowering time. {ECO:0000269|PubMed:17653269}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00257 | DAP | Transfer from AT2G02450 | Download |
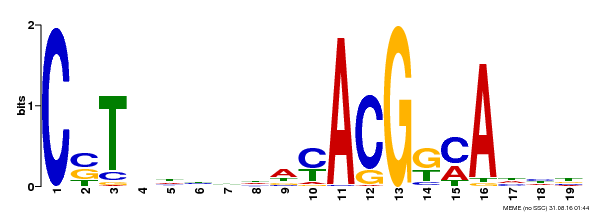
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013607098.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian regulation with a peak of expression at dawn under continuous light conditions (PubMed:17653269). Circadian regulation with a peak of expression around dusk and lowest expression around dawn under continuous light conditions (at protein level) (PubMed:17653269). {ECO:0000269|PubMed:17653269}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC189452 | 0.0 | AC189452.2 Brassica rapa subsp. pekinensis cultivar Inbred line 'Chiifu' clone KBrB072E02, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013607098.1 | 0.0 | PREDICTED: NAC domain-containing protein 55-like isoform X2 | ||||
| Swissprot | Q9ZVP8 | 1e-162 | NAC35_ARATH; NAC domain-containing protein 35 | ||||
| TrEMBL | A0A0D3E7K3 | 0.0 | A0A0D3E7K3_BRAOL; Uncharacterized protein | ||||
| TrEMBL | A0A3N6TUH6 | 0.0 | A0A3N6TUH6_BRACR; Uncharacterized protein | ||||
| STRING | Bo9g071690.1 | 0.0 | (Brassica oleracea) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G02450.2 | 1e-149 | NAC domain containing protein 35 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



