 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013608859.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 336aa MW: 36336.2 Da PI: 8.2909 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 118.2 | 3.3e-37 | 42 | 101 | 4 | 63 |
zf-Dof 4 kalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkksss 63
+al+cprC+s ntkfCyynnysl+qPryfCk C+ryWt+GG+lrn+P+Gg rknk+sss
XP_013608859.1 42 HALNCPRCNSLNTKFCYYNNYSLTQPRYFCKDCKRYWTQGGSLRNIPIGGVVRKNKRSSS 101
6799****************************************************9875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 2.0E-27 | 26 | 91 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 1.7E-32 | 43 | 98 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 28.099 | 44 | 98 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 46 | 82 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 336 aa Download sequence Send to blast |
MDTAKWPQKE FIVKPMNETC LKQPNPAAAP VEEKKARPEK YHALNCPRCN SLNTKFCYYN 60 NYSLTQPRYF CKDCKRYWTQ GGSLRNIPIG GVVRKNKRSS SSSSSNSNSS SSSKKTLSVN 120 NKTPPPLQQL NLKTVENDHV AKACSHGFGN AHEAKDLNSA FSLGFEIGHH HHQSIPEFLQ 180 VVPSSMKNHT PVSTSSALEL LGISNSSTTS SNSSPAFVTY PNVHDSSVYT AASPGFGMSY 240 PQFQEFMRPP LGFSLDGGDS LHLEEGSSGT DNGRPLLPFE SLKLHVSSSS TNSGGGGNGN 300 DHKREKEEGE ADNSVGFWNG MLSAGDSAAS AGGPWQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
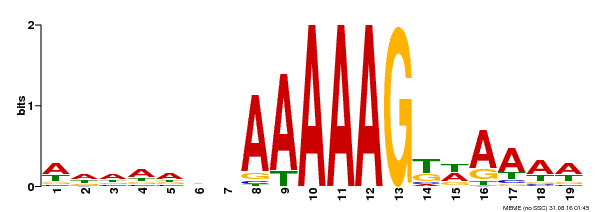
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00210 | DAP | Transfer from AT1G64620 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013608859.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013608859.1 | 0.0 | PREDICTED: dof zinc finger protein DOF1.8 isoform X1 | ||||
| Swissprot | Q84JQ8 | 1e-133 | DOF18_ARATH; Dof zinc finger protein DOF1.8 | ||||
| TrEMBL | A0A0D3E473 | 0.0 | A0A0D3E473_BRAOL; Uncharacterized protein | ||||
| STRING | Bo9g032170.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM14932 | 17 | 20 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G64620.1 | 1e-126 | Dof family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



