 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013609055.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 336aa MW: 37671.8 Da PI: 7.3409 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57 | 4.5e-18 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g W++eEde+l+++++++G g+W+++++ g+ R++k+c++rw +yl
XP_013609055.1 14 KGLWSPEEDEKLLRYITKYGHGCWSSVPKQAGLQRCGKSCRLRWINYL 61
678*******************************************97 PP
| |||||||
| 2 | Myb_DNA-binding | 45.1 | 2.2e-14 | 67 | 110 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg+++++E+ l++++++ lG++ W+ Ia+ ++ gRt++++k+ w++
XP_013609055.1 67 RGAFSQDEENLIIELHAVLGNR-WSQIAAQLP-GRTDNEIKNLWNS 110
89********************.*********.*********9985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-28 | 6 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 25.666 | 9 | 65 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 8.24E-30 | 12 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.1E-13 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.1E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 3.24E-12 | 17 | 61 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.6E-25 | 65 | 116 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.201 | 66 | 116 | IPR017930 | Myb domain |
| SMART | SM00717 | 6.0E-13 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.9E-13 | 67 | 110 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.83E-9 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 336 aa Download sequence Send to blast |
MGRHSCCYKQ KLRKGLWSPE EDEKLLRYIT KYGHGCWSSV PKQAGLQRCG KSCRLRWINY 60 LRPDLKRGAF SQDEENLIIE LHAVLGNRWS QIAAQLPGRT DNEIKNLWNS SLKKKLRLRG 120 IDPVTHKLLA EIETGTDDNT TPVEKCQTTY LIETEGSSST TTGSTNHNNS NTDHLYTGNF 180 GFQRLSLETG SRIQTGIWIP QTGRNHHVDT VPSAVVLPGS MFSSGLTDST TGYRSSNLGL 240 TELDNSFSTG PMITEQHLQE SNYNNSTFFG TGNLSWGLTM EENQFTISNN SLQNHSNSSL 300 YSEIKSETNF FGTEAANVGM WPCNQLQPQQ HAYGHI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 3e-29 | 12 | 116 | 5 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed specifically in guard cells (PubMed:16005292). Expressed in sink tissues, such as xylem, roots and developing seeds (PubMed:22708996). {ECO:0000269|PubMed:16005292, ECO:0000269|PubMed:22708996}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that coordinates a small network of downstream target genes required for several aspects of plant growth and development, such as xylem formation and xylem cell differentiation, and lateral root formation (PubMed:22708996). Regulates a specific set of target genes by binding DNA to the AC cis-element 5'-ACCTAC-3' (PubMed:23741471). Functions as a transcriptional regulator of stomatal closure. Plays a role the regulation of stomatal pore size independently of abscisic acid (ABA) (PubMed:16005292). Required for seed coat mucilage deposition during the development of the seed coat epidermis (PubMed:19401413). Involved in the induction of trichome initiation and branching by positively regulating GL1 and GL2. Required for gibberellin (GA) biosynthesis and degradation by positively affecting the expression of the enzymes that convert GA9 into the bioactive GA4, as well as the enzymes involved in the degradation of GA4 (PubMed:28207974). {ECO:0000269|PubMed:16005292, ECO:0000269|PubMed:19401413, ECO:0000269|PubMed:22708996, ECO:0000269|PubMed:23741471, ECO:0000269|PubMed:28207974}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00427 | DAP | Transfer from AT4G01680 | Download |
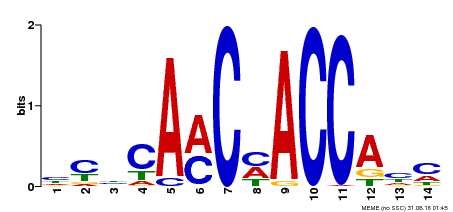
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013609055.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013609055.1 | 0.0 | PREDICTED: transcription repressor MYB6 | ||||
| Refseq | XP_013722795.1 | 0.0 | transcription repressor MYB6 | ||||
| Swissprot | Q8VZQ2 | 2e-84 | MYB61_ARATH; Transcription factor MYB61 | ||||
| TrEMBL | A0A0D3DZW4 | 0.0 | A0A0D3DZW4_BRAOL; Uncharacterized protein | ||||
| STRING | Bo9g004210.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4 | 28 | 2646 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G01680.1 | 0.0 | myb domain protein 55 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



