 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013619674.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 487aa MW: 55271.3 Da PI: 7.5147 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 53.8 | 4.5e-17 | 241 | 287 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT +Ed ll ++v ++G+++W+ Ia++++ gR +kqc++rw+++l
XP_013619674.1 241 KGQWTSDEDRLLMQLVVLHGTKRWSQIAKMLQ-GRVGKQCRERWHNHL 287
799*****************************.*************97 PP
| |||||||
| 2 | Myb_DNA-binding | 54.6 | 2.6e-17 | 294 | 335 | 2 | 45 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ W++eEd +l++a+k+ G++ W+ Iar+++ gRt++ +k++w+
XP_013619674.1 294 DGWSEEEDRILIEAHKEIGNR-WAEIARKLP-GRTENTIKNHWN 335
56*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 29.224 | 236 | 291 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.86E-31 | 238 | 334 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.2E-15 | 240 | 289 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.8E-27 | 242 | 295 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.02E-13 | 244 | 287 | No hit | No description |
| Pfam | PF13921 | 1.3E-17 | 244 | 301 | No hit | No description |
| SMART | SM00717 | 2.7E-16 | 292 | 340 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 20.836 | 292 | 342 | IPR017930 | Myb domain |
| CDD | cd00167 | 5.25E-13 | 296 | 338 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-21 | 296 | 340 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010439 | Biological Process | regulation of glucosinolate biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1904095 | Biological Process | negative regulation of endosperm development | ||||
| GO:2000692 | Biological Process | negative regulation of seed maturation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 487 aa Download sequence Send to blast |
MEFQSVIKMH YPYLASVIFD DNFSLKDVHP SLTDDASCVQ NVHHKPFHAK LDKSHVPSII 60 NPDLCHKNLT LNISNTTFDH IPNLNISPRH YDMQPHIFDT VSKDIYNGIT PSPCTEAFEA 120 YLRGISNDQD LFDMACNTSS QHVEPNDVPN ISHVPRDNTM WENNQNQGII FGTESIFNLA 180 MVDSNLYGAP KPIGILGANE DTIMNQRHNN QIMIKVDQIK KSKRFQMRRG SKLTRKASII 240 KGQWTSDEDR LLMQLVVLHG TKRWSQIAKM LQGRVGKQCR ERWHNHLRPD IKKDGWSEEE 300 DRILIEAHKE IGNRWAEIAR KLPGRTENTI KNHWNATKRK QHSRKSKGKD EVSLALGSNT 360 LQNYIRSVTY NDDTLMTATS NSNGNNGRRN VRGKGKNVIV DASDYNKKND ENECKYIVDG 420 VMNLDLEKST ICVSRSTSAS GSTSGSGSGV TMEVDEPMED SWMVMHGCDE VMMNEIALLE 480 MIARGHV |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1mse_C | 7e-44 | 239 | 341 | 2 | 104 | C-Myb DNA-Binding Domain |
| 1msf_C | 7e-44 | 239 | 341 | 2 | 104 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Expressed in embryos from the early heart stage and throughout embryogenesis (PubMed:18695688, PubMed:19066902). Induced at the onset of the maturation phase in the endosperm, in a high and homogeneous repartition (PubMed:18695688, PubMed:19066902, PubMed:25194028, PubMed:27681170). {ECO:0000269|PubMed:18695688, ECO:0000269|PubMed:19066902, ECO:0000269|PubMed:25194028, ECO:0000269|PubMed:27681170}. | |||||
| Uniprot | TISSUE SPECIFICITY: Mainly expressed in siliques (PubMed:18695688, PubMed:19066902). Also detected at low levels in leaves and flowers (PubMed:19066902). {ECO:0000269|PubMed:18695688, ECO:0000269|PubMed:19066902}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
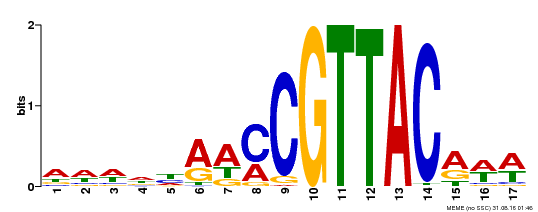
| UniProt | Transcription activator that recognizes the motif 5'-TAACGG-3' in the promoter of endosperm-induced genes (PubMed:27681170, PubMed:25194028, PubMed:19066902). Promotes vegetative-to-embryonic transition and the formation of somatic embryos from root explants in a WUS-independent manner but via the expression of embryonic genes (e.g. LEC1, LEC2, FUS3 and WUS) (PubMed:18695688). May play an important role during embryogenesis and seed maturation (PubMed:19066902, PubMed:25194028). Together with MYB115, activates the transcription of S-ACP-DES2/AAD2 and S-ACP-DES3/AAD3 thus promoting the biosynthesis of omega-7 monounsaturated fatty acid in seed endosperm (PubMed:27681170). Regulates negatively maturation genes in the endosperm (PubMed:25194028). {ECO:0000269|PubMed:18695688, ECO:0000269|PubMed:19066902, ECO:0000269|PubMed:25194028, ECO:0000269|PubMed:27681170}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00385 | DAP | Transfer from AT3G27785 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013619674.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by LEC2. {ECO:0000269|PubMed:25194028}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB018114 | 6e-36 | AB018114.1 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone: MGF10. | |||
| GenBank | AF334817 | 6e-36 | AF334817.2 Arabidopsis thaliana putative transcription factor (MYB118) mRNA, complete cds. | |||
| GenBank | AY550306 | 6e-36 | AY550306.1 Arabidopsis thaliana MYB transcription factor (At3g27785) mRNA, complete cds. | |||
| GenBank | CP002686 | 6e-36 | CP002686.1 Arabidopsis thaliana chromosome 3, complete sequence. | |||
| GenBank | DQ446708 | 6e-36 | DQ446708.1 Arabidopsis thaliana clone pENTR221-At3g27785 myb family transcription factor (At3g27785) mRNA, complete cds. | |||
| GenBank | DQ653111 | 6e-36 | DQ653111.1 Arabidopsis thaliana clone 0000013325_0000009613 unknown mRNA. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013619674.1 | 0.0 | PREDICTED: uncharacterized protein LOC106326201 isoform X1 | ||||
| Swissprot | Q9LVW4 | 0.0 | MY118_ARATH; Transcription factor MYB118 | ||||
| TrEMBL | A0A0D3AWA4 | 0.0 | A0A0D3AWA4_BRAOL; Uncharacterized protein | ||||
| STRING | Bo2g144970.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6052 | 26 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G27785.1 | 0.0 | myb domain protein 118 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



