 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013626711.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 308aa MW: 34194.8 Da PI: 8.1814 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 175.1 | 2e-54 | 20 | 147 | 1 | 128 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpkkvkaeekewyfFskrdkkyatgkrknratksgyWkatgkdkevlsk 95
+ppGfrFhPtdeel+++yL kkv ++++++ +i++vd++k+ePw+Lp+k+k +ekewyfF+ rd+ky+tg r+nrat++gyWkatgkd+e++s+
XP_013626711.1 20 MPPGFRFHPTDEELITYYLLKKVLDSSFSC-AAISQVDLNKSEPWELPEKAKMGEKEWYFFTLRDRKYPTGLRTNRATEAGYWKATGKDREIKSS 113
79****************************.88***************99999****************************************98 PP
NAM 96 .kgelvglkktLvfykgrapkgektdWvmheyrl 128
+++l g+kktLvfykgrapkgek+ Wvmheyrl
XP_013626711.1 114 kTNSLLGMKKTLVFYKGRAPKGEKSCWVMHEYRL 147
56677***************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 4.45E-61 | 16 | 172 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 57.46 | 20 | 172 | IPR003441 | NAC domain |
| Pfam | PF02365 | 1.3E-28 | 21 | 147 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0010014 | Biological Process | meristem initiation | ||||
| GO:0010160 | Biological Process | formation of organ boundary | ||||
| GO:0010223 | Biological Process | secondary shoot formation | ||||
| GO:0048467 | Biological Process | gynoecium development | ||||
| GO:0051782 | Biological Process | negative regulation of cell division | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 308 aa Download sequence Send to blast |
MDMDVFNGWE RSRYEDETVM PPGFRFHPTD EELITYYLLK KVLDSSFSCA AISQVDLNKS 60 EPWELPEKAK MGEKEWYFFT LRDRKYPTGL RTNRATEAGY WKATGKDREI KSSKTNSLLG 120 MKKTLVFYKG RAPKGEKSCW VMHEYRLDGK FSYHYITSSA KDEWVLSKVC LKSSVVSRET 180 KLISSSGGVN CSSSSASAGS LIAPMIDAYA TEHVSCFSNT SAVHADASFP PTYLPAPLPP 240 SLPRQPRRFG DDVAFGQFMD VGASGQFSID AAFLPNLPSL PPTVFTPPSQ PFGVYGGASA 300 VSSWPFAL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3swm_A | 6e-49 | 20 | 178 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_B | 6e-49 | 20 | 178 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_C | 6e-49 | 20 | 178 | 20 | 174 | NAC domain-containing protein 19 |
| 3swm_D | 6e-49 | 20 | 178 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_A | 6e-49 | 20 | 178 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_B | 6e-49 | 20 | 178 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_C | 6e-49 | 20 | 178 | 20 | 174 | NAC domain-containing protein 19 |
| 3swp_D | 6e-49 | 20 | 178 | 20 | 174 | NAC domain-containing protein 19 |
| Search in ModeBase | ||||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: First observed in young embryonic SAM. Later confined to the boundaries between cotyledon primordia and the SAM. In mature embryos, localized around first leaves primordia. Only weakly present in vegetative SAM. In inflorescence, observed at the boundaries between floral organ primordia. In callus, expressed during transition to shoot development, with a progressive restriction to specific areas corresponding to future shoot apex. {ECO:0000269|PubMed:11245578, ECO:0000269|PubMed:12492830}. | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed in inflorescence stems, rosette leaves, aerial parts of seedlings, flowers, floral buds and roots. {ECO:0000269|PubMed:11245578}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator of STM and KNAT6. Involved in molecular mechanisms regulating shoot apical meristem (SAM) formation during embryogenesis and organ separation. Required for the fusion of septa of gynoecia along the length of the ovaries. Activates the shoot formation in callus in a STM-dependent manner. Seems to act as an inhibitor of cell division. {ECO:0000269|PubMed:10079219, ECO:0000269|PubMed:10750709, ECO:0000269|PubMed:11245578, ECO:0000269|PubMed:12163400, ECO:0000269|PubMed:12492830, ECO:0000269|PubMed:12610213, ECO:0000269|PubMed:12787253, ECO:0000269|PubMed:14617069, ECO:0000269|PubMed:15202996, ECO:0000269|PubMed:15294871, ECO:0000269|PubMed:15500463, ECO:0000269|PubMed:15723790, ECO:0000269|PubMed:16798887, ECO:0000269|PubMed:17122068, ECO:0000269|PubMed:17287247, ECO:0000269|PubMed:9212461}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00357 | DAP | Transfer from AT3G15170 | Download |
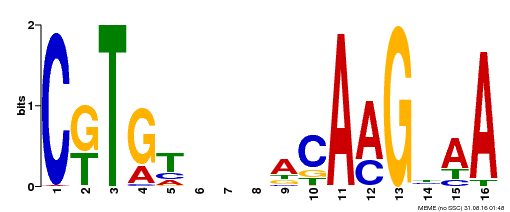
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013626711.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By BRM, at the chromatin level, and conferring a very specific spatial expression pattern. Directly induced by ESR2 in response to cytokinins. Precise spatial regulation by post-transcriptional repression directed by the microRNA miR164. {ECO:0000269|PubMed:15202996, ECO:0000269|PubMed:15294871, ECO:0000269|PubMed:15723790, ECO:0000269|PubMed:16854978, ECO:0000269|PubMed:17056621, ECO:0000269|PubMed:17287247}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | JN169828 | 0.0 | JN169828.1 Brassica oleracea isolate BoCF3 CUC1-like gene, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013626711.1 | 0.0 | PREDICTED: protein CUP-SHAPED COTYLEDON 1-like | ||||
| Swissprot | Q9FRV4 | 1e-158 | NAC54_ARATH; Protein CUP-SHAPED COTYLEDON 1 | ||||
| TrEMBL | A0A0D3BBF7 | 0.0 | A0A0D3BBF7_BRAOL; Uncharacterized protein | ||||
| STRING | Bo3g066690.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13481 | 15 | 28 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G15170.1 | 1e-144 | NAC family protein | ||||



