 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013634080.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 309aa MW: 33150.2 Da PI: 8.9908 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 123.8 | 5.6e-39 | 77 | 136 | 3 | 62 |
zf-Dof 3 ekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
e+alkcprC+stntkfCy+nnysl+qPr+fCk+CrryWt+GGalrnvPvGgg+r+n++++
XP_013634080.1 77 EAALKCPRCESTNTKFCYFNNYSLTQPRHFCKTCRRYWTRGGALRNVPVGGGCRRNRRTK 136
6789*****************************************************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD007478 | 5.0E-35 | 60 | 129 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 3.6E-32 | 79 | 134 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.478 | 80 | 134 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 82 | 118 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 309 aa Download sequence Send to blast |
MIFSSIQAYL DSSNWQQAPP SNYNQGGEGA SATGGHGLGP QLQPQSQPQP NGSGSSGSIR 60 PGSMVDRARH ANVAIPEAAL KCPRCESTNT KFCYFNNYSL TQPRHFCKTC RRYWTRGGAL 120 RNVPVGGGCR RNRRTKSSNN NNNSTATSNN TSFSSAASGN ASTISAILSS NYGGNQESFL 180 SQILSPVRLM NTTYTTDNNM SLLNCGGLNQ DLRSIQMGNS GGSLMSCVDE WRSTSHHQQP 240 QSLGGGNCEA SSNTNPSSNG FYPFESPRIT SALASQFSSV KVEDNPYKWV NVNGNCSSWT 300 DLSTFGSSR |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 120 | 133 | RNVPVGGGCRRNRR |
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | DEVELOPMENTAL STAGE: Specific to the midveins, containing narrow procambial cell files. Expressed in procambial cells of leaf primordia, roots and embryos, prior to the completion of xylem differentiation. {ECO:0000269|PubMed:17583520}. | |||||
| Uniprot | TISSUE SPECIFICITY: Specific to the vascular tissues (PubMed:17583520). The PEAR proteins (e.g. DOF2.4, DOF5.1, DOF3.2, DOF1.1, DOF5.6 and DOF5.3) form a short-range concentration gradient that peaks at protophloem sieve elements (PSE) (PubMed:30626969). {ECO:0000269|PubMed:17583520, ECO:0000269|PubMed:30626969}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Probably involved in early processes for vascular development (PubMed:17583520). The PEAR proteins (e.g. DOF2.4, DOF5.1, DOF3.2, DOF1.1, DOF5.6 and DOF5.3) activate gene expression that promotes radial growth of protophloem sieve elements. Triggers the transcription of HD-ZIP III genes, especially in the central domain of vascular tissue (PubMed:30626969). {ECO:0000250|UniProtKB:Q9M2U1, ECO:0000269|PubMed:17583520, ECO:0000269|PubMed:30626969}. | |||||
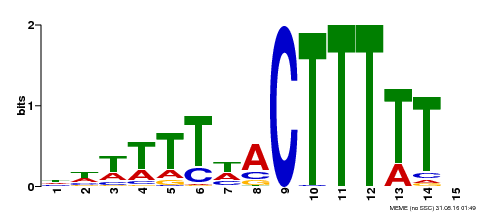
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00295 | DAP | Transfer from AT2G37590 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013634080.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cytokinin in procambium. Antagonized by the HD-ZIP III proteins and by mobile miR165 and miR166 microRNAs. {ECO:0000269|PubMed:30626969}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC189307 | 0.0 | AC189307.2 Brassica rapa subsp. pekinensis cultivar Inbred line 'Chiifu' clone KBrB033O04, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013634080.1 | 0.0 | PREDICTED: dof zinc finger protein DOF2.4-like | ||||
| Refseq | XP_013680574.1 | 0.0 | dof zinc finger protein DOF2.4 | ||||
| Swissprot | O80928 | 1e-154 | DOF24_ARATH; Dof zinc finger protein DOF2.4 | ||||
| TrEMBL | A0A0D3C4U0 | 0.0 | A0A0D3C4U0_BRAOL; Uncharacterized protein | ||||
| TrEMBL | A0A3P6D470 | 0.0 | A0A3P6D470_BRAOL; Uncharacterized protein | ||||
| STRING | Bo4g187060.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM11354 | 17 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G37590.1 | 1e-136 | DNA binding with one finger 2.4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



