 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013634648.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Brassiceae; Brassica
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 448aa MW: 50564.7 Da PI: 5.6281 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 34.5 | 3.8e-11 | 241 | 282 | 8 | 54 |
HHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 8 rErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
Er RR + N++f+ L+ l+P++ +K + a+i+ +A++YIk+L
XP_013634648.1 241 TERERRVHFNDRFFDLKNLIPNP-----TKSDRASIVGEAIDYIKEL 282
59*********************.....9****************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 4.71E-13 | 225 | 295 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 14.126 | 233 | 282 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 9.0E-9 | 239 | 288 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.4E-8 | 241 | 282 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 1.5E-11 | 241 | 294 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 6.32E-11 | 242 | 287 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0048658 | Biological Process | anther wall tapetum development | ||||
| GO:0052543 | Biological Process | callose deposition in cell wall | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 448 aa Download sequence Send to blast |
MEEERGSIYE EMPCFDANTP ASVTAESSFS QVQPPQILVA GSTSNSNCSL DVEEFHLSPQ 60 DCPQASSTPL QFHISPLPPP PRDNTQFNLI HQMSHNQQQQ QHSNWGNGYQ DFNNMCPNST 120 TTPDLLSLLH LPRCSLPLPN SSISFSDIMS SSSAAAVMYD PLFHLNFPLQ SRDNNQLPNG 180 SCLLGVEEQI QMDANGGGIN MMYYEPEHNN NTGFESGAFE FGNGGNNRRG RGSGKSRTFP 240 TERERRVHFN DRFFDLKNLI PNPTKSDRAS IVGEAIDYIK ELLRTIEEFK MLVEKKKFGK 300 FRSKKKPKTC GEEDLEQEQE GDNVSYRPQS EVDQSCFNMK NKNKSLRCSW LKRKSKVTEV 360 DVRIIDDEVT IKVVQKKKIN CLLFTTKVLD QLKLDLHHVA GGQIGEHYSF LFNTKICEES 420 CVYASGIADT VMEVVEKQYM EAVPTNGY |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00285 | DAP | Transfer from AT2G31220 | Download |
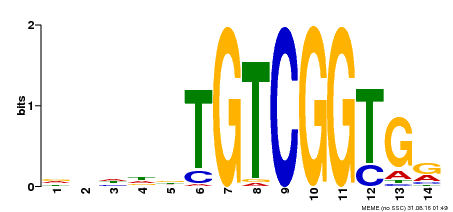
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_013634648.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013634648.1 | 0.0 | PREDICTED: transcription factor bHLH10 | ||||
| Swissprot | Q84TK1 | 0.0 | BH010_ARATH; Transcription factor bHLH10 | ||||
| TrEMBL | A0A078FSY8 | 0.0 | A0A078FSY8_BRANA; BnaC04g42050D protein | ||||
| TrEMBL | A0A0D3C3D5 | 0.0 | A0A0D3C3D5_BRAOL; Uncharacterized protein | ||||
| STRING | Bo4g176070.1 | 0.0 | (Brassica oleracea) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1355 | 26 | 96 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G31220.1 | 1e-174 | bHLH family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



