 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013900848.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Sphaeropleales; Selenastraceae; Monoraphidium
|
||||||||
| Family | C3H | ||||||||
| Protein Properties | Length: 487aa MW: 48506.7 Da PI: 8.4931 | ||||||||
| Description | C3H family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-CCCH | 45.5 | 1.2e-14 | 106 | 131 | 1 | 26 |
--S---SGGGGTS--TTTTT-SS-SS CS
zf-CCCH 1 yktelCrffartGtCkyGdrCkFaHg 26
yktelC+++++ GtC+yG +C+FaHg
XP_013900848.1 106 YKTELCKNWSALGTCRYGVKCQFAHG 131
9************************9 PP
| |||||||
| 2 | zf-CCCH | 36.6 | 7.7e-12 | 145 | 168 | 2 | 25 |
-S---SGGGGTS--TTTTT-SS-S CS
zf-CCCH 2 ktelCrffartGtCkyGdrCkFaH 25
k lCr+fa tG C+yG rC+F H
XP_013900848.1 145 KSDLCRTFAITGSCPYGTRCRFQH 168
5689******************** PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF81995 | 2.35E-6 | 7 | 83 | No hit | No description |
| Gene3D | G3DSA:1.25.10.10 | 2.3E-4 | 11 | 66 | IPR011989 | Armadillo-like helical |
| Gene3D | G3DSA:4.10.1000.10 | 8.3E-16 | 102 | 137 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 2.22E-9 | 104 | 136 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 8.9E-9 | 105 | 132 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 15.76 | 105 | 133 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 6.2E-11 | 106 | 131 | IPR000571 | Zinc finger, CCCH-type |
| Gene3D | G3DSA:4.10.1000.10 | 1.5E-10 | 140 | 170 | IPR000571 | Zinc finger, CCCH-type |
| SuperFamily | SSF90229 | 2.88E-7 | 142 | 170 | IPR000571 | Zinc finger, CCCH-type |
| SMART | SM00356 | 2.7E-5 | 143 | 170 | IPR000571 | Zinc finger, CCCH-type |
| PROSITE profile | PS50103 | 14.69 | 143 | 171 | IPR000571 | Zinc finger, CCCH-type |
| Pfam | PF00642 | 1.1E-8 | 145 | 168 | IPR000571 | Zinc finger, CCCH-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 487 aa Download sequence Send to blast |
MSSVFAQPEQ QSQQHDAVNQ HQQHQNYQQQ QQQQEQQQQQ QQQQQQQQLQ QQQQQLQQQH 60 TLANLPMQQA QHSTPRREAA PPLLLSPQRS ASGGVDPYCK PNNFLYKTEL CKNWSALGTC 120 RYGVKCQFAH GTLELRPASR PPRAKSDLCR TFAITGSCPY GTRCRFQHAE PGSASAAAAL 180 AAARASPQQG GSAGAGAAAA LGARRQAEAA RRLAAAGGEY CAGGLFDASG AECPSPLPNM 240 LALSGAASAH RAGGGAMLPM LPQASPAAAS AGAAAAAAGL YSPASAGGGR IGGRSSFGSG 300 AGGSSGSFDA PLSGAGSGPL VNPLAAWDAA AAGGGAGHVG TTTQGSMGGG SARVFAQPLG 360 AIGTPSGTGT ALMQQGGIPG FAICSSPGAT LWGAAGIAGQ APSPLAAAAV PTPSLQPMPQ 420 QQAAFGGSGA AEPPAGPGLL ASGDPIQDLL QALTLGQGQS QGQCQLGEGP RGSAAASPPA 480 DAPRTNA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1rgo_A | 1e-21 | 106 | 168 | 4 | 66 | Butyrate response factor 2 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00383 | ampDAP | Transfer from AT1G68200 | Download |
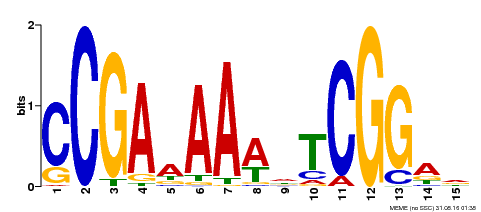
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013900848.1 | 0.0 | Zinc finger protein zfs1 | ||||
| TrEMBL | A0A0D2L3Q6 | 0.0 | A0A0D2L3Q6_9CHLO; Zinc finger protein zfs1 | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP3623 | 13 | 14 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G68200.1 | 3e-19 | C3H family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 25739010 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



