 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_013906551.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Sphaeropleales; Selenastraceae; Monoraphidium
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 620aa MW: 63200.4 Da PI: 6.7546 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 23.8 | 9.4e-08 | 191 | 240 | 5 | 54 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkk 54
kr +r NR++A rs +R+ +++ eLe kv++L++ +L +ele+ +
XP_013906551.1 191 KRVKRILANRLSAARSKERRVKYTLELEAKVQALDGDLARLSNELESTRA 240
8999**************************************99997665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 2.0E-6 | 187 | 251 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.082 | 189 | 239 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 3.5E-7 | 191 | 242 | No hit | No description |
| Pfam | PF00170 | 6.6E-7 | 191 | 238 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 1.37E-7 | 191 | 240 | No hit | No description |
| CDD | cd14703 | 2.14E-14 | 192 | 237 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007231 | Biological Process | osmosensory signaling pathway | ||||
| GO:0008272 | Biological Process | sulfate transport | ||||
| GO:0009294 | Biological Process | DNA mediated transformation | ||||
| GO:0009652 | Biological Process | thigmotropism | ||||
| GO:0009970 | Biological Process | cellular response to sulfate starvation | ||||
| GO:0045596 | Biological Process | negative regulation of cell differentiation | ||||
| GO:0051170 | Biological Process | nuclear import | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0003682 | Molecular Function | chromatin binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0043621 | Molecular Function | protein self-association | ||||
| GO:0051019 | Molecular Function | mitogen-activated protein kinase binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 620 aa Download sequence Send to blast |
MVAAGPLYVG GGDDALKPAA PFPGGADQQL QQLQQVQPQP FQLLQQHQQA DLVGGRPART 60 RKPTDKVALG VAVAGKRSHA GCKGKDPATV AAAAAVALER TGGETGSGCH QRRNSFTPAL 120 PSVLERDGGE DAGGMESPPR DEIGEEDEED DDSDNYSGSG RGKKGGGRRG GANGGGASGR 180 NRGELLTLDP KRVKRILANR LSAARSKERR VKYTLELEAK VQALDGDLAR LSNELESTRA 240 RAAAAQRAQA EADGRALGLQ QVLAHASAAN QALAAEVREL QRCLGLPEQL PSLTAPSLAV 300 AAPGASPPQS TQAQQDQHQQ QPVQQEQQSG QTALVIKQQQ PALLGDEDGD VEMLAELLGV 360 TGSPLPHERS GAASGVAPLS PASANTSAGA HYNHHQPHQQ LQVQGWLASM SGHSVPSSPP 420 PTGIAGRGGL HPPRPLLATL SEPLGERIAI PDEASTTPSW SGMCTTPAGA AGRASNIAAL 480 PPCLQAQPLL QQQPGLPHEP SGSQPLAFGA APQPQRPSGS QHQRSASAGS ISPLASDAGA 540 LVYGSGMAAT SFVQLKPGAV VFGAAPLQLP QQADTGGRFM TVTTPGPAGT PSTNITAGAA 600 LAPEALLGPG AVPLAPFGGL |
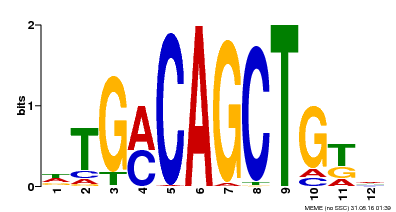
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00184 | DAP | Transfer from AT1G43700 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_013906551.1 | 0.0 | hypothetical protein MNEG_0421 | ||||
| TrEMBL | A0A0D2N5H2 | 0.0 | A0A0D2N5H2_9CHLO; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP6785 | 4 | 4 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G43700.1 | 8e-13 | VIRE2-interacting protein 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Entrez Gene | 25726539 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



