 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015870030.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 577aa MW: 61758.8 Da PI: 9.2645 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 26.7 | 9.7e-09 | 373 | 420 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR++i +++ L++l+P + +k Ka +L + ++Y++sLq
XP_015870030.1 373 SHSLAERVRREKISERMKFLQDLVPGC----NKVTGKAVMLDEIINYVQSLQ 420
8*************************9....677*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 6.58E-11 | 367 | 424 | No hit | No description |
| SuperFamily | SSF47459 | 7.2E-17 | 367 | 436 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 15.663 | 369 | 419 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 7.7E-17 | 369 | 435 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 8.9E-6 | 373 | 420 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.5E-10 | 375 | 425 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 577 aa Download sequence Send to blast |
MVKIAYVARV SNGVGLKANL PSWKTNGFGD PFESALSSIV SSPAASNATA AISGGNNVGG 60 DCVMIRELIG RLGSICNSGE ISPQSCINNN NSNSTNNSCY GTPLNSPPKL NLSMVDPQIR 120 VNLPIPGGTH LPPSSSSSSH HTSLVAPFSA DPGFAERAAR FSSFGSKNFG GLNGQLLGLN 180 ETELPFRSIL RVESGKLSRA SSSNPLSVAG SQMGVQESNK SSPLEGNSAP DKKFNRLSRS 240 STPENAEFGD SREGSSVSEQ IPSGELSVKG ETDANTRKRK SIPRGKTKET PLSPSAKGSK 300 VAAGNDESNA KRSKTEEASG NAKDAAKTKA EANGGSKAAG DGNQKQTKDN SKPPEPPKDY 360 IHVRARRGQA TDSHSLAERV RREKISERMK FLQDLVPGCN KVTGKAVMLD EIINYVQSLQ 420 RQVEFLSMKL ATVNPRSDLN MEALLSKDIF QSRGSLSQNP LYPLDSSVPA FPYGFNPTQM 480 PPLHSNMSNG TETQFPLNNL NSALHRNSSM QLPAIDGFGE AAQQVPKIFE DDLQSVVQMG 540 FGQIQQQSFH GMQNIHEKIQ YLKSESEKLN DFSSNES |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 275 | 280 | TRKRKS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00660 | PBM | Transfer from Pp3c6_9520 | Download |
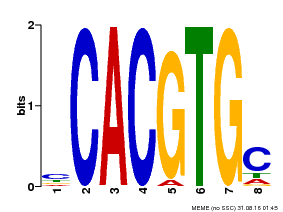
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015870030.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015870030.1 | 0.0 | transcription factor bHLH78-like | ||||
| TrEMBL | A0A0U2RCR8 | 0.0 | A0A0U2RCR8_9ROSA; BHLH transcription factor | ||||
| STRING | EMJ23017 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2530 | 34 | 80 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G07340.1 | 1e-84 | bHLH family protein | ||||



