 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015872823.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 285aa MW: 31160.8 Da PI: 8.4722 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 33.1 | 9.7e-11 | 167 | 214 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
+h+ +Er RR++i +++ L++l+P + +k Ka +L + ++YI+sLq
XP_015872823.1 167 SHSLAERARREKISERMKILQDLVPGC----NKVIGKALVLDEIINYIQSLQ 214
8*************************9....677*****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 1.14E-17 | 161 | 230 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.32E-9 | 161 | 218 | No hit | No description |
| PROSITE profile | PS50888 | 15.828 | 163 | 213 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 6.9E-18 | 163 | 230 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 4.2E-8 | 167 | 214 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.0E-10 | 169 | 219 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0048446 | Biological Process | petal morphogenesis | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 285 aa Download sequence Send to blast |
MDPSTLINEG SLSNGNAASY NLSEIWQFPM NSGGGESGIV LGLRRGQFAQ TLAQFGDMNR 60 DVSGNDPMSL DQRENHGGGG GGGARKRRDA EEESAKGVST SNGNGNGVSN GDGKKLKISG 120 GRDENRESKT EGEANSGKPG EQNSRPHDPS KQDYIHVRAR RGQATDSHSL AERARREKIS 180 ERMKILQDLV PGCNKVIGKA LVLDEIINYI QSLQRQVEFL SMKLEAVNSR NPGIEVFTSK 240 DVSSSNMHHS LWNLFVPGIA TFHLPNKISL MLSYLRMTLM FIFEL |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the control of petal size, by interfering with postmitotic cell expansion to limit final petal cell size. {ECO:0000269|PubMed:16902407}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00207 | DAP | Transfer from AT1G59640 | Download |
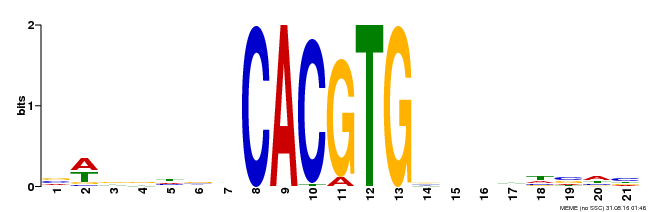
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015872823.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Isoform 1 is up-regulated by PI/AP3, SEP2, SEP3 and AP1, but repressed by AG. {ECO:0000269|PubMed:16902407}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FQ380741 | 8e-59 | FQ380741.1 Vitis vinifera clone SS0ACG35YM08. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015872823.2 | 0.0 | transcription factor BHLH089-like, partial | ||||
| Swissprot | Q0JXE7 | 7e-75 | BPE_ARATH; Transcription factor BPE | ||||
| TrEMBL | A0A2P5D2C5 | 1e-110 | A0A2P5D2C5_PARAD; Basic helix-loop-helix transcription factor | ||||
| STRING | XP_010098235.1 | 1e-110 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1171 | 34 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G59640.2 | 9e-68 | BIG PETAL P | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



