 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015877428.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 376aa MW: 42195.6 Da PI: 8.6156 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 91 | 9.9e-29 | 68 | 121 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH +Fv+ave+LGG+++AtPk +l+lm++kgL+++hvk LQ+YR+
XP_015877428.1 68 PRLRWTPDLHLCFVHAVERLGGQDRATPKLVLQLMNIKGLSIAHVKKALQMYRS 121
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-26 | 64 | 122 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 9.85 | 64 | 124 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.17E-12 | 65 | 122 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.3E-18 | 68 | 123 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.4E-6 | 69 | 120 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 376 aa Download sequence Send to blast |
MVRSDDSSEV SRPSPSNENE PEDDYDQQLD NDSQRLKNGG SSSNSTAEEN AKTGASGSVR 60 QYVRSKTPRL RWTPDLHLCF VHAVERLGGQ DRATPKLVLQ LMNIKGLSIA HVKKALQMYR 120 SKKIEDPNQV LSEQGFFMEG GDHHIYNFSQ LPMLQSFNQW PSNSALRYGD SYRRGHDHQI 180 YFPYSGRAAL GSSRHGLYGS AAERILRNNN IATSSSSNLS PLNNPSFNKI ATWRRNQAQD 240 EYLSFHRSWQ EPVRPNSSSM EANLHHQAIS LKNNGSITTN SRTIQEAGQN PLKRKSSESD 300 CDLDLNLSLK IRPKDDELVV DEKALEGHEE VLSSTLSLSL SSTSSLKLSR VKEKGGDDQD 360 RKQARRMATS TFDLTL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 3e-15 | 69 | 123 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 3e-15 | 69 | 123 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 2e-15 | 69 | 123 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-15 | 69 | 123 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-15 | 69 | 123 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-15 | 69 | 123 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 3e-15 | 69 | 123 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 3e-15 | 69 | 123 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 3e-15 | 69 | 123 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 3e-15 | 69 | 123 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 3e-15 | 69 | 123 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 3e-15 | 69 | 123 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00301 | DAP | Transfer from AT2G40260 | Download |
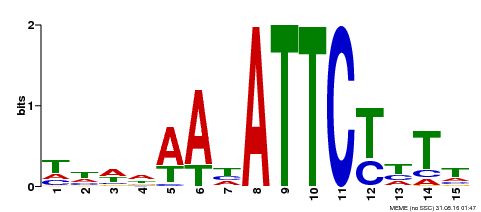
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015877428.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015877428.1 | 0.0 | putative Myb family transcription factor At1g14600 | ||||
| TrEMBL | W9SFW5 | 1e-109 | W9SFW5_9ROSA; Putative Myb family transcription factor | ||||
| STRING | XP_010105426.1 | 1e-110 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2378 | 34 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40260.1 | 1e-52 | G2-like family protein | ||||



