 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015879796.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 395aa MW: 45305 Da PI: 6.2704 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 84.6 | 1.3e-26 | 280 | 367 | 1 | 86 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkr..tsessstcpyfdqle 86
rW++ ev+aLi++r ++e++++ + +k Wee+s+ m g++r++k+Ckekwen+nk++k+ +++ek+r ++++ss pyf++le
XP_015879796.1 280 RWPEGEVQALIAVRASLEHKFQITGSKRSIWEEISIGMCNMGYNRTAKKCKEKWENINKYFKRSAASEKNRdfANGKSSGRPYFHDLE 367
8*********************************************************************966699**********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 0.0042 | 277 | 339 | IPR001005 | SANT/Myb domain |
| Pfam | PF13837 | 4.5E-18 | 279 | 367 | No hit | No description |
| CDD | cd12203 | 3.89E-26 | 279 | 343 | No hit | No description |
| PROSITE profile | PS50090 | 6.922 | 280 | 337 | IPR017877 | Myb-like domain |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 395 aa Download sequence Send to blast |
MEVFSGDRQI PNPDDFPLHI APFPEPTDIL YAHPTALIHS SGITDHPISL PQKLRPIRCN 60 VRSSVSGDLE KGLLLSDEVC SINRELELEV KVETSGEAPN GVGLGDPVQV GSGTVAEGSD 120 PNGRERVVED ETSSPSDDSS DDYSTWSLEE PTNRKRSRRR RRKSRRKLEA FLENLVMKVM 180 EKQEQMHKQL IEMIEKREKE RIIREEAWMQ QEMERLKRDE EARAQETSRN LALISLVQNL 240 MGQEIQIPQI QIPQVQPVVS PGTDDSHQKE MKCCDPNNKR WPEGEVQALI AVRASLEHKF 300 QITGSKRSIW EEISIGMCNM GYNRTAKKCK EKWENINKYF KRSAASEKNR DFANGKSSGR 360 PYFHDLELLY KNGFVHPRNY FSITNNENEA KNLYD |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 153 | 166 | RKRSRRRRRKSRRK |
| 2 | 153 | 167 | RKRSRRRRRKSRRKL |
| 3 | 154 | 167 | KRSRRRRRKSRRKL |
| 4 | 157 | 165 | RRRRRKSRR |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00552 | DAP | Transfer from AT5G47660 | Download |
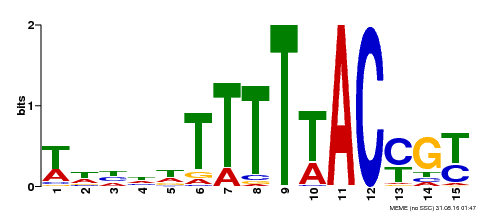
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015879796.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015879796.1 | 0.0 | trihelix transcription factor GT-2 | ||||
| TrEMBL | A0A2P5EXN0 | 1e-126 | A0A2P5EXN0_TREOI; Octamer-binding transcription factor | ||||
| STRING | XP_010087663.1 | 1e-119 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11112 | 33 | 38 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G47660.1 | 2e-33 | Trihelix family protein | ||||



