 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015885201.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 1059aa MW: 116390 Da PI: 5.755 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.3 | 7.2e-18 | 37 | 83 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT+eEde l +av+++ g++Wk+Ia+++ Rt+ qc +rwqk+l
XP_015885201.1 37 KGQWTPEEDEVLRRAVQRFKGKNWKKIAECFK-DRTDVQCLHRWQKVL 83
799****************************9.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 64.5 | 2e-20 | 89 | 135 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+W++eEde+++++v+++G++ W+tI+++++ gR +kqc++rw+++l
XP_015885201.1 89 KGPWSKEEDEIIIELVNKYGPKKWSTISQHLP-GRIGKQCRERWHNHL 135
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 49.5 | 9.9e-16 | 141 | 183 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT+eE++ l++a++ +G++ W+ + ++ gRt++++k++w+
XP_015885201.1 141 KEAWTQEEELALIRAHQIYGNK-WAELTKFLP-GRTDNSIKNHWN 183
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.782 | 32 | 83 | IPR017930 | Myb domain |
| SMART | SM00717 | 8.6E-15 | 36 | 85 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.6E-15 | 37 | 83 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.2E-22 | 38 | 90 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 9.33E-16 | 38 | 93 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.04E-13 | 40 | 83 | No hit | No description |
| PROSITE profile | PS51294 | 32.985 | 84 | 139 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.36E-31 | 86 | 182 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.6E-19 | 88 | 137 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.2E-19 | 89 | 135 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.24E-17 | 91 | 135 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 5.8E-28 | 91 | 139 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 19.674 | 140 | 190 | IPR017930 | Myb domain |
| SMART | SM00717 | 3.2E-15 | 140 | 188 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.9E-21 | 140 | 190 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 7.3E-14 | 141 | 184 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 7.25E-12 | 143 | 183 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1059 aa Download sequence Send to blast |
MEGDRSISTP SDGLGDGFQK IRALHGRTSG PTRRSTKGQW TPEEDEVLRR AVQRFKGKNW 60 KKIAECFKDR TDVQCLHRWQ KVLNPELVKG PWSKEEDEII IELVNKYGPK KWSTISQHLP 120 GRIGKQCRER WHNHLNPAIN KEAWTQEEEL ALIRAHQIYG NKWAELTKFL PGRTDNSIKN 180 HWNSSVKKKL DSYLKSGLLA QFQGLPHVGH QNQPMVSSSS RMQSSGDDSG HKGTEAEEIS 240 ECSQDSTFSG RFPSSNDMAS VVLCTRKEFQ GTNNSGLGKD PNPSSASCSV PYYPSVEGSA 300 FSVPEISPEI GSAAKFLEQG FPHDAETSIS GDIQFNLDEL PNISSLELAC ERSGIPTHCL 360 GSDVRQGENV QFQTSEGLSV STSMGTMPLS SNKPAHMLIS DDECCTVLFS EAMNNRCFSS 420 KTLAKGSDFV GLGGCTGSLL SHSSNIPMSE ASGTAATQLY CPSNSNATGT SCSQTFHPTV 480 ISANDRPLIF GGESNHLFGT QEYEYIISSD DGFVFANDCA NSPCNDGTDA IGLHEQSDTL 540 KDSSKLVPVN TFSSRSDTQT CPMDRRPDEL REQKDTGALC YEPPRFPSLD VPFFSCDLVQ 600 SSSDMQQEYS PLGIRQLMMS SMNCLTPFRL WDSPTRDNSP DAVLKSAAKT FTGTPTILKK 660 RHRELFSPLS DRRCDKKLDT HVTSSLMRDF SRLDVMFDDS GSHKAPSVSP SCCQDGNTRN 720 SAASAADKEN QGHLTPERLK KGSDSTEISD GSIPEKDVED SESQEKTKHG VVDVDAKTKI 780 DAGPTSEIEE MPSEILVEHN INDLLLYSPD QVNLKAERAL GSGMKTPRNQ YHKSSEPTSN 840 QCVGQTSAER QCASVCSPSI SGKKLVSCSV AETCVRSDSS LILPEMMGCN AGSDATTETI 900 SMFGSTPFKR SIESPSAWKS PWFINSFLPC PRIDTEITIE DFGYFMSPGN RSYDAIGLMK 960 QVSEQTAAAY ANAQEILGDE TPETLFQRRT SHERRDQENN HVRPNQLQSH SHLPQNILTE 1020 RRTLDFSECG TPGKGIEHGK SSTSVSFSSP SSYLLKGCR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 2e-69 | 37 | 190 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 2e-69 | 37 | 190 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00466 | DAP | Transfer from AT4G32730 | Download |
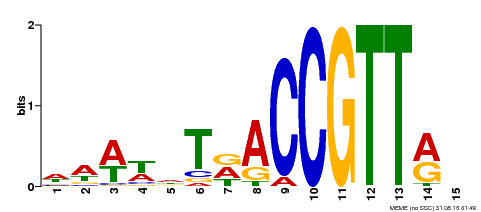
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015885201.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015885201.1 | 0.0 | transcription factor MYB3R-1-like | ||||
| TrEMBL | A0A251RDM5 | 0.0 | A0A251RDM5_PRUPE; Uncharacterized protein | ||||
| STRING | EMJ26596 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3727 | 33 | 62 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G32730.2 | 0.0 | Homeodomain-like protein | ||||



