 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015886299.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1066aa MW: 119329 Da PI: 5.4324 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 185.6 | 5.2e-58 | 21 | 136 | 3 | 118 |
CG-1 3 kekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrr 97
+ ++rwl++ ei++iL n++k+++t+e++trp+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+++vl+cyYah+e+n++fqrr
XP_015886299.1 21 EAQHRWLRPAEICEILRNYQKFRITSEPPTRPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSIDVLHCYYAHGEDNENFQRR 115
449******************************************************************************************** PP
CG-1 98 cywlLeeelekivlvhylevk 118
+yw+Le+el +iv+vhylevk
XP_015886299.1 116 SYWMLEQELMHIVFVHYLEVK 136
******************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 85.308 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.2E-84 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.5E-51 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 3.5E-5 | 454 | 565 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 1.7E-4 | 480 | 555 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 3.67E-14 | 480 | 565 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 2.8E-18 | 668 | 775 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 19.571 | 668 | 775 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 4.15E-16 | 670 | 773 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 3.6E-18 | 672 | 779 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 6.9E-8 | 675 | 740 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 670 | 681 | 710 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.511 | 681 | 713 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 9.2E-4 | 714 | 743 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 10.472 | 714 | 746 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 5.84E-8 | 888 | 940 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.076 | 889 | 911 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.766 | 890 | 919 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0019 | 891 | 910 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.017 | 912 | 934 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.304 | 913 | 937 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.7E-4 | 915 | 934 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1066 aa Download sequence Send to blast |
MAERGSYGLS PRLDIQQLSV EAQHRWLRPA EICEILRNYQ KFRITSEPPT RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHEK LKVGSIDVLH CYYAHGEDNE NFQRRSYWML 120 EQELMHIVFV HYLEVKGNRT NIGGVRETDE FSQDLSKASP QISSSSANNC KEPSGNTDYT 180 SPSSSLTSLY EDADTGDSHQ ASSTYHSYSG SPQPQVGNGL LMNNIDVSFL TGPAAKNHEG 240 HSSINGADYM PHLEKDRPMY NDSPGYIVGQ QKTLGLGSWE EILEQCTMGF HTDPACTGVA 300 LEQQNMTLNG LSACDSTAEK ELDSSLPTHS SWQIPFEDNS LLLPKLSVDQ PPNSELPCYQ 360 DDLFNTLLHQ QKEKPAQSNL QAQLPNAESE CLIASNADDD IYADGNMNYS LSLRRQLFDG 420 EEGLKKVDSF SRWVSKELGE VDNLQMQSSS GISWSTVECG NVVDDSSLSP SLSQDQLFSI 480 IDVSPKWGYA DLETEVLTTG TFLKSQQEVA KYNWSCMFGE VEGPAEILVD GVLCCHAPPH 540 RSGQVPFYVT CSNRIACSEV REFDYLLGST KGLDITNIYG SDTDEMLLHV RLERLLSLGS 600 VKPSKNIFKD VTEKKDLINK IISLKEEEES NQKVDLTPEN DLSQYEVKEH LLKKLIKDKL 660 YSWLFHKVTE DGKGPSVLDD NGQGALHLAA ALGYNWAIKP IVTSGVSINF RDVNGWTALH 720 WAALYGREQT VALLVSLGAA PGAVTDPSPE FPLGRTPADL ASVTGQKGIS GFLAESSLTS 780 YLSSLTVNEP KNDCAAENSG TTAVQTVSER TATPMNYFDM PDTLSLKDSL TAVRNATQAA 840 DRIHLMFRMQ SFERRQLSDL GDDEFGLSDE HALSLLATKT SKAGPSYGQA HAAAVHIQKK 900 YRGWKKRKEF LIIRQRIVKI QAHVRGHQVR KQYRAIIWSV GILEKIILRW RRKGSGLRGF 960 RNDAVIKDSQ PDPQHMPLKD DDYDFFKEGR KQTEERLQSA LTRVKSMVQY PEGRAQYRRL 1020 LNVVDGFRET KVDDMALNSE TADSDENLVD IDTLLHDDTF MSIAFE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
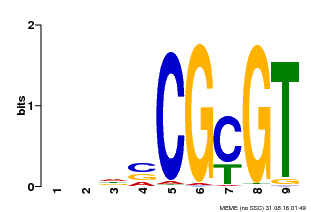
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015886299.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015886299.1 | 0.0 | calmodulin-binding transcription activator 2-like isoform X1 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A2P5E8N4 | 0.0 | A0A2P5E8N4_TREOI; Notch | ||||
| STRING | XP_008237378.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7406 | 26 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



