 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015890633.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1069aa MW: 120343 Da PI: 5.5852 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 179.2 | 4.9e-56 | 16 | 132 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqr 96
++ ++rwl++ ei++iL n+++++++ e+++ p+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+he+LK g+++vl+cyYah+e+n++fqr
XP_015890633.1 16 MEAQHRWLRPAEICEILRNYKRFRIAPEPANMPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHERLKAGSIDVLHCYYAHGEDNENFQR 110
5569******************************************************************************************* PP
CG-1 97 rcywlLeeelekivlvhylevk 118
r+yw+Lee+l++ivlvhy+evk
XP_015890633.1 111 RSYWMLEEDLSHIVLVHYREVK 132
*******************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 82.891 | 11 | 137 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.7E-79 | 14 | 132 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 3.5E-49 | 17 | 130 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 1.7E-6 | 485 | 571 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 3.0E-8 | 486 | 570 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 5.91E-19 | 486 | 571 | IPR014756 | Immunoglobulin E-set |
| PROSITE profile | PS50297 | 17.29 | 651 | 791 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.95E-15 | 670 | 781 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 3.32E-11 | 678 | 779 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.4E-15 | 680 | 785 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 0.032 | 719 | 748 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 9.217 | 719 | 751 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 2200 | 759 | 788 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.12E-7 | 887 | 945 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 16 | 894 | 916 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.144 | 896 | 924 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.066 | 897 | 915 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0039 | 917 | 939 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.487 | 918 | 942 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.2E-4 | 920 | 939 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1069 aa Download sequence Send to blast |
MAESRRYGLD IEQILMEAQH RWLRPAEICE ILRNYKRFRI APEPANMPPS GSLFLFDRKV 60 LRYFRKDGHN WRKKKDGKTV KEAHERLKAG SIDVLHCYYA HGEDNENFQR RSYWMLEEDL 120 SHIVLVHYRE VKGNRTSFNR IRENEDAETA PNSEIDSSFS SSFPPNSYQI SQTTDTTSLN 180 SAQASEYEDA ESAYNQASST LHSFLELQRP MAEQINSGLS DPYYPMMFSN DYQGKSSAIP 240 GIDISSLPQT DINEGSKSVG VTYEPRKNLD FPLRKNILVN TSAGTQSLPL QPSLSAIQSE 300 NLGIVQKQEQ ENFGQLFSEG IGQRLEFGSQ PQVQEEWQAS GGHSSSLSKW PADQNLHQDA 360 ASNLASERET NGVELLQSQH PNTQHEYDLK SVQENNVFLE GKPNYISGIK QSLLDSSFTD 420 EGLKKLDSFN RWMSKELGDV NESHMQTSSE AYWDTVEAEN ADGDSSQVRL DTYMLGPSLS 480 QDQLFTIIDF SPNWAFEDSE VKVLITGRFL DHQAESSKWS CMFGEVEVPA EVIADGVLRC 540 HTPIHKAGRV PFYVTCSNRL ACSEVREFEY RVNEVRDMDL KYDDSSCTTE ELNLRFGKLL 600 CLDSACPTSG PNNLVEKSQL SSKISLLLRE DEDEWDQMLK LTSENNFSVE RVEEQLHQKL 660 LKGKLHGWLL QKVAEGGKGA SVLDEGGQGV LHFAAALDYE WALEPTIIAG VSVNFRDVNG 720 WTALHWAAFC GRERTVASLI SLGAAPGALT DPSPKYQTGG KTPSDLAYAK GHKGIAGYLA 780 ESALSAHLLS LNLDKKEGNA AETSGVKAVH TISERVATPV KDGDLNDRLS LKDSLAAVCN 840 ATQAAARIHQ VFRVQSFQRK QLKEYGDDKF GMSDEQALSL IAVKSAKQGH HDEHVNAAAI 900 RIQNKFRSWK GRKDFLIIRQ RIVKIQAHVR GHQVRKNYRK ITWSVGIVEK IILRWRRKGS 960 GLRGFKSEAL TEGPSKENSL SKEDDDDFLK EGRKQAEVRL QKALNRVKSM VQYPEARDQY 1020 RRLLNVVSEF QGTKVQFDTD PNNSETADFD DDLIDLEALL DEDTYMPTA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cxk_A | 1e-14 | 487 | 571 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_B | 1e-14 | 487 | 571 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_C | 1e-14 | 487 | 571 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_D | 1e-14 | 487 | 571 | 9 | 89 | calmodulin binding transcription activator 1 |
| 2cxk_E | 1e-14 | 487 | 571 | 9 | 89 | calmodulin binding transcription activator 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3'. Binds calmodulin in a calcium-dependent manner in vitro (PubMed:12218065). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance in association with CAMTA1 and CAMTA2 (PubMed:23581962). Required for the cold-induced expression of DREB1B/CBF1, DREB1C/CBF2, ZAT12 and GOLS3 (PubMed:19270186). Involved in response to cold. Contributes together with CAMTA5 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). Involved together with CAMTA2 and CAMTA4 in the positive regulation of a general stress response (GSR) (PubMed:25039701). Involved in the regulation of GSR amplitude downstream of MEKK1 (PubMed:25157030). Involved in the regulation of a set of genes involved in defense responses against pathogens (PubMed:18298954). Involved in the regulation of both basal resistance and systemic acquired resistance (SAR) (PubMed:21900483). Acts as negative regulator of plant immunity (PubMed:19122675, PubMed:21900483, PubMed:22345509, PubMed:28407487). Binds to the promoter of the defense-related gene EDS1 and represses its expression (PubMed:19122675). Binds to the promoter of the defense-related gene NDR1 and represses its expression (PubMed:22345509). Involved in defense against insects (PubMed:23072934, PubMed:22371088). Required for tolerance to the generalist herbivore Trichoplusia ni, and contributes to the positive regulation of genes associated with glucosinolate metabolism (PubMed:23072934). Required for tolerance to Bradysia impatiens larvae. Mediates herbivore-induced wound response (PubMed:22371088). Required for wound-induced jasmonate accumulation (PubMed:23072934, PubMed:22371088). Involved in the regulation of ethylene-induced senescence by binding to the promoter of the senescence-inducer gene EIN3 and repressing its expression (PubMed:22345509). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:18298954, ECO:0000269|PubMed:19122675, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:21900483, ECO:0000269|PubMed:22345509, ECO:0000269|PubMed:22371088, ECO:0000269|PubMed:23072934, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25157030, ECO:0000269|PubMed:28351986, ECO:0000269|PubMed:28407487, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00042 | PBM | Transfer from AT2G22300 | Download |
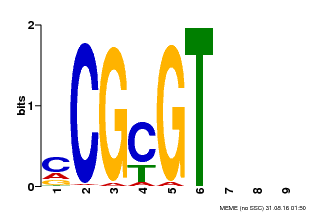
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015890633.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, salt, wounding, ethylene and methyl jasmonate (PubMed:11162426, PubMed:12218065). Induced by infection with the fungal pathogen Golovinomyces cichoracearum (powdery mildew) and the bacterial pathogen Pseudomonas syringae pv tomato strain DC3000 (PubMed:22345509). {ECO:0000269|PubMed:11162426, ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:22345509}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AM434540 | 6e-38 | AM434540.2 Vitis vinifera contig VV78X210461.8, whole genome shotgun sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015890633.1 | 0.0 | calmodulin-binding transcription activator 3 isoform X2 | ||||
| Swissprot | Q8GSA7 | 0.0 | CMTA3_ARATH; Calmodulin-binding transcription activator 3 | ||||
| TrEMBL | A0A2P5EKP9 | 0.0 | A0A2P5EKP9_TREOI; Notch | ||||
| STRING | XP_010087599.1 | 0.0 | (Morus notabilis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G22300.2 | 0.0 | signal responsive 1 | ||||



