 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015891267.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 289aa MW: 33214.8 Da PI: 5.8887 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 57.5 | 3e-18 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT eEd++l+++++++G g+W++ ++ g+ R++k+c++rw +yl
XP_015891267.1 14 KGPWTSEEDQILINYIQRHGHGNWRALPKLAGLLRCGKSCRLRWTNYL 61
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 56.3 | 7.4e-18 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++++eE+e +++++++lG++ W+tIa++++ gRt++++k+ w+++l
XP_015891267.1 67 RGNFSPEEEETIIKLHEMLGNR-WSTIASRLP-GRTDNEIKNVWHTHL 112
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 3.9E-26 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 16.91 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 9.87E-32 | 10 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.9E-14 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.4E-16 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.01E-10 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 26.309 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.3E-27 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.6E-15 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 4.0E-16 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.69E-10 | 69 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 289 aa Download sequence Send to blast |
MVRAPCCEKM GLKKGPWTSE EDQILINYIQ RHGHGNWRAL PKLAGLLRCG KSCRLRWTNY 60 LRPDIKRGNF SPEEEETIIK LHEMLGNRWS TIASRLPGRT DNEIKNVWHT HLKKRLRHNH 120 NTEETKRRRS TANIISKQNQ EAKKEEDSTM INYNNNFSSN VIKTKSESIE YNRPVSPPQS 180 SSEISSVTTG TTDSNNNNND DFSMNVENFS EIMDENFWSE VLSSAENSDE VVNDFSAMGN 240 VEQELQFLNS PIASMEPVHG SNSYDSGMDF WYNIFTRAGE TSEFLLPDL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 2e-27 | 12 | 116 | 5 | 108 | B-MYB |
| 1h8a_C | 4e-27 | 12 | 116 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 112 | 128 | KKRLRHNHNTEETKRRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in cold-regulation of CBF genes and in the development of freezing tolerance. May be part of a complex network of transcription factors controlling the expression of CBF genes and other genes in response to cold stress. Binds to the MYB recognition sequences in the promoters of CBF1, CBF2 and CBF3 genes (PubMed:17015446). Involved in drought and salt tolerance. May enhance expression levels of genes involved in abscisic acid (ABA) biosynthesis and signaling, as well as those encoding stress-protective proteins (PubMed:19161942). {ECO:0000269|PubMed:17015446, ECO:0000269|PubMed:19161942}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00375 | DAP | Transfer from AT3G23250 | Download |
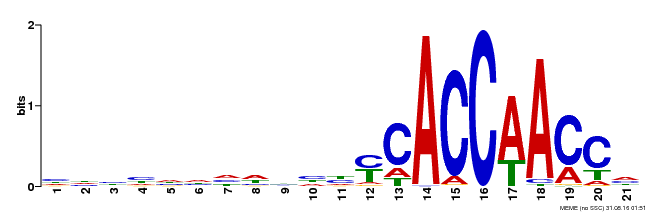
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015891267.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by abscisic acid (ABA) and drought stress (PubMed:19161942). Induced by salt stress (PubMed:19161942). {ECO:0000269|PubMed:19161942}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015891267.1 | 0.0 | transcription factor MYB13 | ||||
| Swissprot | Q9LTC4 | 4e-86 | MYB15_ARATH; Transcription factor MYB15 | ||||
| TrEMBL | A0A2I4F6P9 | 1e-108 | A0A2I4F6P9_JUGRE; myb-related protein Myb4-like | ||||
| TrEMBL | B9H8L9 | 1e-108 | B9H8L9_POPTR; Uncharacterized protein | ||||
| STRING | POPTR_0005s24550.1 | 1e-109 | (Populus trichocarpa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G23250.1 | 1e-82 | myb domain protein 15 | ||||



