 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015891841.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 418aa MW: 46922 Da PI: 4.5088 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.3 | 2.7e-16 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+grWT eEd++l+++++ G g+W++ ++ g+ R++k+c++rw +yl
XP_015891841.1 14 KGRWTSEEDQILIKYIQVNGEGSWRSLPKNAGLLRCGKSCRLRWINYL 61
79******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 54 | 3.9e-17 | 67 | 112 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+ + eE++l+v++++ lG++ W++Ia++++ gRt++++k++w+++l
XP_015891841.1 67 RGNISSEEEDLIVKLHASLGNR-WSLIASHLP-GRTDNEIKNYWNSHL 112
7899******************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 9.1E-22 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 17.053 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.49E-28 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 7.4E-12 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.9E-14 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.90E-8 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 25.779 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.4E-25 | 65 | 116 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.8E-16 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.9E-15 | 67 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 1.86E-10 | 71 | 112 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0009813 | Biological Process | flavonoid biosynthetic process | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 418 aa Download sequence Send to blast |
MGRAPCCEKV GLKKGRWTSE EDQILIKYIQ VNGEGSWRSL PKNAGLLRCG KSCRLRWINY 60 LRADLKRGNI SSEEEDLIVK LHASLGNRWS LIASHLPGRT DNEIKNYWNS HLSRKIHTFR 120 KPINIGNESL PSIVDLAKIN VNPPTKRKGG RTSRWAMKKN KSYVQKEPVI SSSTEINVAD 180 LEINEVVPLP QTPAIDNESL SVSNTDECMV WNPDSEDKHN HHHHNNNNKK VPDEEAGGGE 240 TENSSILCLD VEKEKNINEI SGPYDEEQGI DDGMLCFNDI MDSELLEANG VYMTINEEKE 300 SIDAVGLSEE RESRVTFQND KMGMSDQEIE SSGNLSSSNG GGGIGEVGDW YSCSSMNSGL 360 DDHVGLDFWD WESVVQEQDI DENEKMLSWL WESDSSETLK FGDVDSEKQS AMVAWLLS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 3e-24 | 12 | 116 | 2 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| 1h8a_C | 3e-24 | 12 | 116 | 25 | 128 | MYB TRANSFORMING PROTEIN |
| 1mse_C | 3e-24 | 12 | 116 | 2 | 105 | C-Myb DNA-Binding Domain |
| 1msf_C | 3e-24 | 12 | 116 | 2 | 105 | C-Myb DNA-Binding Domain |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00627 | PBM | Transfer from PK06182.1 | Download |
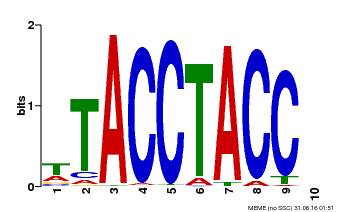
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015891841.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015891841.1 | 0.0 | transcription factor MYB30 | ||||
| TrEMBL | A0A2I4EP80 | 1e-131 | A0A2I4EP80_JUGRE; transcription factor MYB12-like | ||||
| STRING | EOX95226 | 1e-128 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF31 | 34 | 817 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47460.1 | 3e-78 | myb domain protein 12 | ||||



