 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015894425.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 405aa MW: 46834.2 Da PI: 6.6346 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 88.2 | 9.6e-28 | 90 | 172 | 2 | 86 |
trihelix 2 WtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqle 86
W ++e++ Li +rrem+ ++++k++k+lWe++s+kmre+gf rsp++C++kw+nl k++kk k++++++ s +++y++++e
XP_015894425.1 90 WVQDETRSLIGLRREMDGLFNTSKSNKHLWEQISAKMREKGFDRSPTMCTDKWRNLLKEFKKAKHQDRGG--SGSAKMSYYKEIE 172
********************************************************************98..34558*****998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF81995 | 9.68E-9 | 32 | 67 | No hit | No description |
| PROSITE profile | PS50090 | 8.455 | 82 | 146 | IPR017877 | Myb-like domain |
| Gene3D | G3DSA:1.10.10.60 | 4.6E-4 | 83 | 148 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 0.0022 | 86 | 148 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 1.78E-28 | 88 | 153 | No hit | No description |
| Pfam | PF13837 | 2.8E-20 | 89 | 174 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 405 aa Download sequence Send to blast |
MYLSEKPRPI DFYKEEGSRD MMIEVVSNGE IPQPHHHPQQ QTQQQQQQQH QQQQQQQQQQ 60 QQAHQHQMLL GDSSGEDHEV KAPKKRAETW VQDETRSLIG LRREMDGLFN TSKSNKHLWE 120 QISAKMREKG FDRSPTMCTD KWRNLLKEFK KAKHQDRGGS GSAKMSYYKE IEEILRERSK 180 TAQYKSPTPP KVDSYMQFSD KALEDTSISF GPVEAPWCAA SGRPTLNLER RLDHDGHPLA 240 ITTADAVAAS GVPPWNWRET PGNGGESQSY AGRVISVKWG DYTRRIGIDA TAEAIKDAIK 300 SAFGLRTRRA FWLEDEYQII RSLDRDMPLG NYILHVDEGL AIKVCLYDES DHIPVHTEEK 360 IFYTEDDYRD FLTRRAWTCL REFDGYRNID NMDDLRPGAI YRGVS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2jmw_A | 3e-49 | 84 | 170 | 1 | 86 | DNA binding protein GT-1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
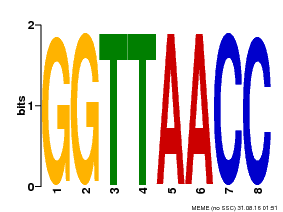
| UniProt | Probable transcription factor that binds specifically to the core DNA sequence 5'-GGTTAA-3'. May act as a molecular switch in response to light signals. {ECO:0000269|PubMed:10437822, ECO:0000269|PubMed:15044016, ECO:0000269|PubMed:7866025}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00004 | PBM | Transfer from 920224 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015894425.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015894425.1 | 0.0 | trihelix transcription factor GT-1 isoform X1 | ||||
| Swissprot | Q9FX53 | 0.0 | TGT1_ARATH; Trihelix transcription factor GT-1 | ||||
| TrEMBL | A0A314ZGR1 | 0.0 | A0A314ZGR1_PRUYE; Trihelix transcription factor GT-1-like isoform X2 | ||||
| STRING | XP_008222025.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6817 | 32 | 48 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G13450.1 | 0.0 | Trihelix family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



