 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015895705.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | RAV | ||||||||
| Protein Properties | Length: 389aa MW: 42945.1 Da PI: 9.3894 | ||||||||
| Description | RAV family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 43.4 | 8.6e-14 | 79 | 127 | 1 | 55 |
AP2 1 sgykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
s+ykGV + +grW A+I++ ++r++lg+f ++eAaka++ a+++++g
XP_015895705.1 79 SKYKGVVPQP-NGRWGAQIYEK-----HQRVWLGTFNEEDEAAKAYDVAAQRFRG 127
89****9888.8*********3.....5**********99*************98 PP
| |||||||
| 2 | B3 | 103 | 1.6e-32 | 219 | 318 | 1 | 96 |
EEEE-..-HHHHTT-EE--HHH.HTT....---..--SEEEEEETTS-EEEEEE..EEETTEEEE-TTHHHHHHHHT--TT-EEEEEE-SSSEE. CS
B3 1 ffkvltpsdvlksgrlvlpkkfaeeh....ggkkeesktltledesgrsWevkliyrkksgryvltkGWkeFvkangLkegDfvvFkldgrsefe 91
f+k+ tpsdv+k++rlv+pk++ae+h +g++ +++ l +ed g++W+++++y+++s++yvltkGW++Fvk+++Lk+gD+v F+++ s+++
XP_015895705.1 219 FEKAVTPSDVGKLNRLVIPKQHAEKHfplqSGSTSKGVLLNFEDVGGKVWRFRYSYWNSSQSYVLTKGWSRFVKEKNLKAGDIVSFQRSTGSDKQ 313
89****************************888899***************************************************88768888 PP
.EEEE CS
B3 92 lvvkv 96
l++ +
XP_015895705.1 314 LYIDW 318
88877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 3.8E-9 | 79 | 127 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.39E-25 | 79 | 134 | No hit | No description |
| SuperFamily | SSF54171 | 4.32E-17 | 79 | 135 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 1.8E-29 | 80 | 141 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 2.4E-20 | 80 | 135 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 20.231 | 80 | 135 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:2.40.330.10 | 7.7E-43 | 212 | 330 | IPR015300 | DNA-binding pseudobarrel domain |
| SuperFamily | SSF101936 | 9.42E-31 | 217 | 321 | IPR015300 | DNA-binding pseudobarrel domain |
| CDD | cd10017 | 1.17E-28 | 218 | 307 | No hit | No description |
| Pfam | PF02362 | 7.7E-30 | 219 | 319 | IPR003340 | B3 DNA binding domain |
| SMART | SM01019 | 2.0E-25 | 219 | 322 | IPR003340 | B3 DNA binding domain |
| PROSITE profile | PS50863 | 14.607 | 219 | 322 | IPR003340 | B3 DNA binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0048573 | Biological Process | photoperiodism, flowering | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 389 aa Download sequence Send to blast |
MEGSCTDEST TSDSISISPS TTPPPLPFLT HPLPPPPPIK TTTTTKSPPE SLCRVGSGTS 60 VILDSENGVE AESRKLPSSK YKGVVPQPNG RWGAQIYEKH QRVWLGTFNE EDEAAKAYDV 120 AAQRFRGRDA VTNFKPLSSE TDGDDEIEAA FLNSHSKAEI VDMLRKHTYN DELEQSKRNY 180 GLDNNNNNNS PHKKSGNFRE IGSALGSSDR VQKAREQLFE KAVTPSDVGK LNRLVIPKQH 240 AEKHFPLQSG STSKGVLLNF EDVGGKVWRF RYSYWNSSQS YVLTKGWSRF VKEKNLKAGD 300 IVSFQRSTGS DKQLYIDWKT RSTSSGGASN QTAVGPVQPV QMVRLFGVNI FKIPGSDGIG 360 GGCNVGKRMR EMELFELECS KKPRIIGAL |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wid_A | 6e-60 | 216 | 329 | 11 | 125 | DNA-binding protein RAV1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional repressor of flowering time on long day plants. Acts directly on FT expression by binding 5'-CAACA-3' and 5'-CACCTG-3 sequences. Functionally redundant with TEM2. {ECO:0000269|PubMed:18718758}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00024 | PBM | Transfer from AT1G25560 | Download |
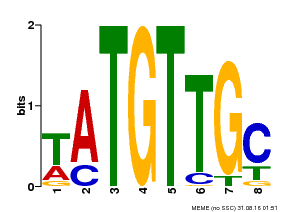
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015895705.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Expressed with a circadian rhythm showing a peak at dawn. {ECO:0000269|PubMed:18718758}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015895705.1 | 0.0 | AP2/ERF and B3 domain-containing transcription factor RAV1-like isoform X2 | ||||
| Swissprot | Q9C6M5 | 1e-152 | RAVL1_ARATH; AP2/ERF and B3 domain-containing transcription repressor TEM1 | ||||
| TrEMBL | A0A2C9UJZ4 | 0.0 | A0A2C9UJZ4_MANES; Uncharacterized protein | ||||
| STRING | cassava4.1_010157m | 0.0 | (Manihot esculenta) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G25560.1 | 1e-148 | RAV family protein | ||||



