 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015896991.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 589aa MW: 64872.8 Da PI: 7.1531 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 49.2 | 1.2e-15 | 59 | 105 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g WT++Ede l av+ + g++Wk+Ia+ ++ R++ qc +rwqk+l
XP_015896991.1 59 KGGWTPQEDETLRTAVAAFKGKSWKKIAEFFP-DRSEVQCLHRWQKVL 105
688*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 62.5 | 8.3e-20 | 111 | 157 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEd+++v++v ++G+ W++Ia+ ++ gR +kqc++rw+++l
XP_015896991.1 111 KGPWTQEEDDKIVELVSKYGPTKWSLIAKSLP-GRIGKQCRERWHNHL 157
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 54.3 | 3.1e-17 | 163 | 206 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+++WT eE++ l +a++ +G++ W+ Ia+ ++ gRt++ +k++w++
XP_015896991.1 163 KDAWTLEEELALMNAHRTHGNK-WAEIAKVLP-GRTDNAIKNHWNS 206
679*******************.*********.***********96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 14.758 | 54 | 105 | IPR017930 | Myb domain |
| SMART | SM00717 | 7.8E-14 | 58 | 107 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.6E-14 | 59 | 105 | IPR001005 | SANT/Myb domain |
| SuperFamily | SSF46689 | 1.29E-14 | 60 | 115 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-21 | 61 | 117 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.33E-12 | 62 | 105 | No hit | No description |
| PROSITE profile | PS51294 | 33.439 | 106 | 161 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 6.36E-32 | 108 | 204 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.9E-18 | 110 | 159 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 7.1E-19 | 111 | 157 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.32E-16 | 113 | 157 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-27 | 118 | 164 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.8E-16 | 162 | 210 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 22.208 | 162 | 212 | IPR017930 | Myb domain |
| Pfam | PF00249 | 1.4E-15 | 163 | 206 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.0E-23 | 165 | 212 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.61E-12 | 165 | 208 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 589 aa Download sequence Send to blast |
MAEEKLEECC LENKQSTAAS SSSVSEGSGS AILKSPGICS PASTSPTHRR TSGPIRRAKG 60 GWTPQEDETL RTAVAAFKGK SWKKIAEFFP DRSEVQCLHR WQKVLNPDLV KGPWTQEEDD 120 KIVELVSKYG PTKWSLIAKS LPGRIGKQCR ERWHNHLNPE IKKDAWTLEE ELALMNAHRT 180 HGNKWAEIAK VLPGRTDNAI KNHWNSSLKK KLDFYLATGK LPPVAKSSPQ NGPKDTGRFT 240 ATKKLVVSSN KELDSVSQTS LGNADTYQLE EDGQDQLESS APPHDMGASS SVHQNESADS 300 EGVECKPGTS SMDPSCSNSE SIPKCKNYGI NSKHKIENST IDIGPKSDIF GINSKPMFEN 360 FGFAGKINED RIIGSPFPFE TPIEGSLYYE PPRFEDSDIS DTHFLQQEYN SSPFLSPVGF 420 FTPPCVKSNG SRMPSPESFL KKAAMSFPNT PSILRKRKIK AKVPFSPSKI EKADGESVRD 480 VCHSFNEHEI EVMDMCHTSD EQEIEKNSCE MPCSQDGNLC ASPSCNGNST IGPNGKGFNA 540 SPPYRLRSKR TAVIKSVEKQ LALKFDKECD GKGSSVAEDC SHATKMEVT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 2e-69 | 58 | 212 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 2e-69 | 58 | 212 | 5 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (By similarity). Transcription repressor that regulates organ growth. Binds to the promoters of G2/M-specific genes and to E2F target genes to prevent their expression in post-mitotic cells and to restrict the time window of their expression in proliferating cells (PubMed:26069325). {ECO:0000250|UniProtKB:Q94FL9, ECO:0000269|PubMed:26069325}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00482 | DAP | Transfer from AT5G02320 | Download |
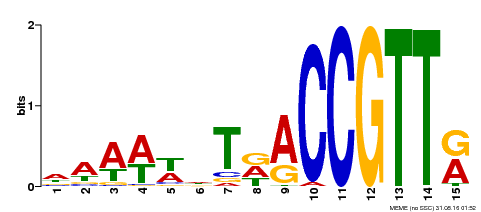
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015896991.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by ethylene and salicylic acid (SA). {ECO:0000269|PubMed:16463103}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015896991.1 | 0.0 | transcription factor MYB3R-3 isoform X1 | ||||
| Swissprot | Q6R032 | 1e-164 | MB3R5_ARATH; Transcription factor MYB3R-5 | ||||
| TrEMBL | A0A2N9HV10 | 0.0 | A0A2N9HV10_FAGSY; Uncharacterized protein | ||||
| STRING | EMJ09642 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6616 | 34 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G02320.2 | 1e-154 | myb domain protein 3r-5 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



