 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015897601.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 397aa MW: 44686.9 Da PI: 9.1564 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.9 | 4e-32 | 70 | 123 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtp+LH rFv+ave+LGG+e+AtPk +l+lm++kgL+++hvkSHLQ+YR+
XP_015897601.1 70 PRLRWTPDLHLRFVHAVERLGGQERATPKLVLQLMNIKGLSIAHVKSHLQMYRS 123
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.791 | 66 | 126 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-29 | 66 | 124 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 8.6E-15 | 69 | 125 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.1E-22 | 70 | 125 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.6E-9 | 71 | 122 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 397 aa Download sequence Send to blast |
MEASLSSGRS ECLKTSPSNQ NDDQGGISES GENDDESNKP QNGGSSSNST IDESEKRGSS 60 VRPYVRSKLP RLRWTPDLHL RFVHAVERLG GQERATPKLV LQLMNIKGLS IAHVKSHLQM 120 YRSKKTDDPS QVIADHRHLV ECGDRNIYNF SQLPMLQGYN YNQTHSSSFR YGYHDNASPW 180 NAYNGNLKHS RSSSSSSSPF YGTDKIFGGN TTTSNWASTF HFPTGISSFS TTHDQQSSSW 240 KIHKPKDEML IQPYQNRESW THYNNLQPKA VQELTGLNKN NNNNNNNNNN NALPDSNTRT 300 SIQDWKMPKR KASDNCNLDL DLSLRITSRN EMDQNHQKSS TLLGEDEVDS NLSLSLYSSS 360 TSLSLKSAKL SRLKVEGTED RKDQHARKRA STLDLTI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 8e-18 | 71 | 125 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 8e-18 | 71 | 125 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 8e-18 | 71 | 125 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 8e-18 | 71 | 125 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 8e-18 | 71 | 125 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 8e-18 | 71 | 125 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 8e-18 | 71 | 125 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 8e-18 | 71 | 125 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 8e-18 | 71 | 125 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 8e-18 | 71 | 125 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 8e-18 | 71 | 125 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 8e-18 | 71 | 125 | 4 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00297 | DAP | Transfer from AT2G38300 | Download |
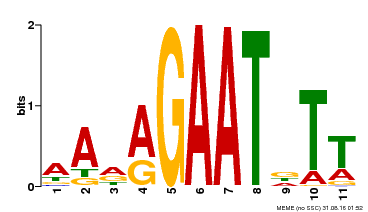
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015897601.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015897601.1 | 0.0 | myb-like protein H | ||||
| TrEMBL | A0A2N9FI59 | 1e-103 | A0A2N9FI59_FAGSY; Uncharacterized protein | ||||
| STRING | XP_010092813.1 | 1e-103 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2378 | 34 | 82 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G38300.1 | 7e-64 | G2-like family protein | ||||



