 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015900006.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 894aa MW: 100860 Da PI: 7.4266 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 168.9 | 7.8e-53 | 3 | 118 | 3 | 118 |
CG-1 3 kekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrr 97
+ ++rwl+++ei+aiL n++++++ ++ + pksg++iL++rk++r+frkDG++wkkkkdgktv+E+he+LKvg e +++yYah+++nptf rr
XP_015900006.1 3 EARSRWLRPNEIHAILCNYKRFTINVKPVNLPKSGTIILFDRKMLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGDEERIHVYYAHGQDNPTFVRR 97
459******************************************************************************************** PP
CG-1 98 cywlLeeelekivlvhylevk 118
cywlL+++le+ivlvhy+e++
XP_015900006.1 98 CYWLLDKSLEHIVLVHYRETQ 118
******************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 78.299 | 1 | 123 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.2E-77 | 1 | 118 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.1E-46 | 3 | 116 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 3.29E-15 | 345 | 432 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 3.9E-4 | 346 | 431 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 3.11E-18 | 531 | 643 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 1.4E-8 | 532 | 610 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.25E-15 | 532 | 640 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 6.9E-18 | 534 | 644 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 17.237 | 539 | 652 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 4.3E-6 | 581 | 610 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.915 | 581 | 613 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 5.84E-7 | 692 | 795 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 33 | 725 | 747 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.327 | 729 | 755 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.272 | 745 | 774 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 270 | 748 | 766 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0011 | 767 | 789 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.768 | 768 | 792 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.1E-4 | 769 | 789 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 10 | 847 | 869 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.279 | 849 | 877 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 894 aa Download sequence Send to blast |
MEEARSRWLR PNEIHAILCN YKRFTINVKP VNLPKSGTII LFDRKMLRNF RKDGHNWKKK 60 KDGKTVKEAH EHLKVGDEER IHVYYAHGQD NPTFVRRCYW LLDKSLEHIV LVHYRETQEL 120 QGSPVTPVNS NSSSASDPSA PWPLSEELDS GTNHAYYAGE NEILVSSDNL TVRNHEQRLH 180 DINTLEWDEL LAIHDPNNSV ASRDKVSFFN QQNQVAGNGL LHGGATSLSP EILSFNILTN 240 PTATSGDIGY NLPQSAYVPT VGAQLNSNVQ RRDSIGAGAG GSLDVLVNDG LHSQDSFGRW 300 INDFITDSSD SVDGSVLETS IPSAQDSFSS LAMHLQSPVS EQIFNITDVS PAWAYSNEKT 360 KILLTGFFRE EYQHLSKSDL LCVCGDISVT AEIVQVGVYR CLVSPHSPGL VNLYISLEGF 420 KPISQVLNFE YRTPALSDQI VSSEERDRWE EFQMQMRLAY LLFSTSKSLE ILTSKASPNA 480 LKEAKKYAQK TSHVSNSWAI FIKSIEDSKI PFSQAKDSLF KLILRNRLKD WLLERVVYGS 540 KISEFDAQGQ GVIHLCAILG YTWAISLFSA SGLSLDFRDK HGWTALHWAA YFGREKMVAV 600 LLSAGAKPNL VTDPTSDNPG GRTAADLASL NGYDGLAGYL SEKALVEQFK DMSIAGNVSG 660 TLDTSTNDFV NPENLCEEEL NLKETLAAYR TAADAAARIQ VAFREHSLKI RTQAVENSNP 720 EIEARNIVAA MKIQHAFRNY ESRKKMAAAA RIQHRFRTWK IRKEFLNLRR QAIKIQAAFR 780 GYQVRRQYRK ILWSVGVLEK AILRWRLKRR GFRGLQVDPI EAVDDQFQGS DTEEDFYKAS 840 KKQAEERVER AVVSVQAMFR SKKAQEEYRR MKMAHNQAML EYEGFLDPEN DLVG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
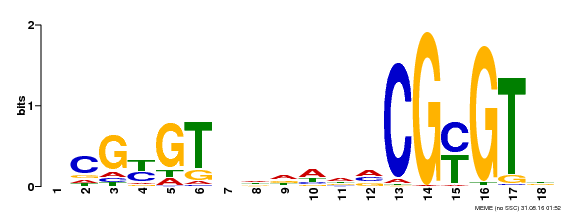
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015900006.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015900006.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X1 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A2P5F618 | 0.0 | A0A2P5F618_TREOI; Notch | ||||
| STRING | XP_009369951.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4288 | 34 | 59 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



