 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015900007.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 892aa MW: 100619 Da PI: 7.4266 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 168.9 | 7.7e-53 | 3 | 118 | 3 | 118 |
CG-1 3 kekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrr 97
+ ++rwl+++ei+aiL n++++++ ++ + pksg++iL++rk++r+frkDG++wkkkkdgktv+E+he+LKvg e +++yYah+++nptf rr
XP_015900007.1 3 EARSRWLRPNEIHAILCNYKRFTINVKPVNLPKSGTIILFDRKMLRNFRKDGHNWKKKKDGKTVKEAHEHLKVGDEERIHVYYAHGQDNPTFVRR 97
459******************************************************************************************** PP
CG-1 98 cywlLeeelekivlvhylevk 118
cywlL+++le+ivlvhy+e++
XP_015900007.1 98 CYWLLDKSLEHIVLVHYRETQ 118
******************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 78.502 | 1 | 123 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.2E-77 | 1 | 118 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 2.1E-46 | 3 | 116 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 3.29E-15 | 343 | 430 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 3.9E-4 | 344 | 429 | IPR002909 | IPT domain |
| SuperFamily | SSF48403 | 3.11E-18 | 529 | 641 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 1.4E-8 | 530 | 608 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.24E-15 | 530 | 638 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 6.8E-18 | 532 | 642 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 17.237 | 537 | 650 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 11.915 | 579 | 611 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 4.3E-6 | 579 | 608 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 5.84E-7 | 690 | 793 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 33 | 723 | 745 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.327 | 727 | 753 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.272 | 743 | 772 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 270 | 746 | 764 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0011 | 765 | 787 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.768 | 766 | 790 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.1E-4 | 767 | 787 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 10 | 845 | 867 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.279 | 847 | 875 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 892 aa Download sequence Send to blast |
MEEARSRWLR PNEIHAILCN YKRFTINVKP VNLPKSGTII LFDRKMLRNF RKDGHNWKKK 60 KDGKTVKEAH EHLKVGDEER IHVYYAHGQD NPTFVRRCYW LLDKSLEHIV LVHYRETQEG 120 SPVTPVNSNS SSASDPSAPW PLSEELDSGT NHAYYAGENE ILVSSDNLTV RNHEQRLHDI 180 NTLEWDELLA IHDPNNSVAS RDKVSFFNQQ NQVAGNGLLH GGATSLSPEI LSFNILTNPT 240 ATSGDIGYNL PQSAYVPTVG AQLNSNVQRR DSIGAGAGGS LDVLVNDGLH SQDSFGRWIN 300 DFITDSSDSV DGSVLETSIP SAQDSFSSLA MHLQSPVSEQ IFNITDVSPA WAYSNEKTKI 360 LLTGFFREEY QHLSKSDLLC VCGDISVTAE IVQVGVYRCL VSPHSPGLVN LYISLEGFKP 420 ISQVLNFEYR TPALSDQIVS SEERDRWEEF QMQMRLAYLL FSTSKSLEIL TSKASPNALK 480 EAKKYAQKTS HVSNSWAIFI KSIEDSKIPF SQAKDSLFKL ILRNRLKDWL LERVVYGSKI 540 SEFDAQGQGV IHLCAILGYT WAISLFSASG LSLDFRDKHG WTALHWAAYF GREKMVAVLL 600 SAGAKPNLVT DPTSDNPGGR TAADLASLNG YDGLAGYLSE KALVEQFKDM SIAGNVSGTL 660 DTSTNDFVNP ENLCEEELNL KETLAAYRTA ADAAARIQVA FREHSLKIRT QAVENSNPEI 720 EARNIVAAMK IQHAFRNYES RKKMAAAARI QHRFRTWKIR KEFLNLRRQA IKIQAAFRGY 780 QVRRQYRKIL WSVGVLEKAI LRWRLKRRGF RGLQVDPIEA VDDQFQGSDT EEDFYKASKK 840 QAEERVERAV VSVQAMFRSK KAQEEYRRMK MAHNQAMLEY EGFLDPENDL VG |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
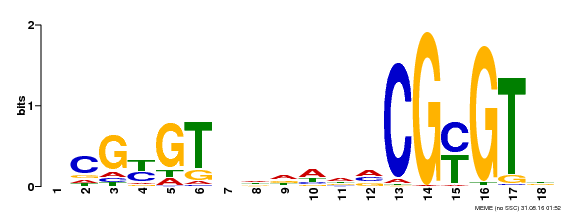
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015900007.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015900007.1 | 0.0 | calmodulin-binding transcription activator 5 isoform X2 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A2P5F618 | 0.0 | A0A2P5F618_TREOI; Notch | ||||
| STRING | XP_009369951.1 | 0.0 | (Pyrus x bretschneideri) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



