 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015902348.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 482aa MW: 53257.4 Da PI: 7.313 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 106.8 | 1.2e-33 | 46 | 100 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprl+Wtp+LHerF+eav+qLGG++kAtPkt+++lm+++gLtl+h+kSHLQkYRl
XP_015902348.1 46 KPRLKWTPDLHERFIEAVNQLGGADKATPKTVMKLMGIPGLTLYHLKSHLQKYRL 100
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 12.174 | 43 | 103 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.2E-31 | 44 | 101 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.18E-16 | 45 | 101 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 5.1E-23 | 46 | 101 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 6.0E-10 | 48 | 99 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 1.4E-24 | 148 | 194 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 482 aa Download sequence Send to blast |
MYHHQQHQVK NIQGSSRMGM PPERHLFLQG GNGPGDSGLV LSTDAKPRLK WTPDLHERFI 60 EAVNQLGGAD KATPKTVMKL MGIPGLTLYH LKSHLQKYRL SKSLHGQANI GAAGKIGTVA 120 VAADRISEAT GPHLNNLSIG PQSNKGLHLS EALQMQIEVQ RRLHEQLEVQ RHLQLRIEAQ 180 GKYLQSVLEK AQETLGRQNL GTVGLEAAKV QLSELVSKVS TQCLNSAFAE LKELQGVCPQ 240 QQTLLQTTQP PDCSMDSCLT CDGSTQKDQE FHNTTSGMVP QLRPSNNDQI NHINAEDHHL 300 MLQKSGTHDQ LPKWCHDHED PKETNMLIGK DIIADQRRSR MLFPVERSSS TTATGLSISI 360 HGGEKTAGSG GSNNGYCDIG RMKGRDTTED DNFLERISSS SSSGRVAVDQ LLHHHHHNNH 420 HQQQIKDHHH EQGVSHHQGY RSSLPNYFAA AAAAKLDLNA HDHENDSSSS CKQFDLNGFS 480 WN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 1e-20 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 1e-20 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 1e-20 | 46 | 102 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 1e-20 | 46 | 102 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 1e-20 | 46 | 102 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 1e-20 | 46 | 102 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 1e-20 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 1e-20 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 1e-20 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 1e-20 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 1e-20 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 1e-20 | 45 | 102 | 1 | 58 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that may activate the transcription of specific genes involved in nitrogen uptake or assimilation (PubMed:15592750). Acts redundantly with MYR1 as a repressor of flowering and organ elongation under decreased light intensity (PubMed:21255164). Represses gibberellic acid (GA)-dependent responses and affects levels of bioactive GA (PubMed:21255164). {ECO:0000269|PubMed:21255164, ECO:0000305|PubMed:15592750}. | |||||
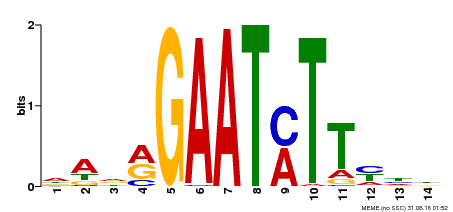
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00331 | DAP | Transfer from AT3G04030 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015902348.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by nitrogen deficiency. {ECO:0000269|PubMed:15592750}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015902348.1 | 0.0 | myb-related protein 2 isoform X1 | ||||
| Swissprot | Q9SQQ9 | 1e-129 | PHL9_ARATH; Myb-related protein 2 | ||||
| TrEMBL | A0A2I4FMC5 | 1e-180 | A0A2I4FMC5_JUGRE; myb-related protein 2-like | ||||
| STRING | EOY11198 | 1e-177 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3852 | 31 | 63 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G04030.3 | 1e-124 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



