 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_015902737.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rhamnaceae; Paliureae; Ziziphus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 211aa MW: 23707.9 Da PI: 10.2754 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 46.9 | 6.3e-15 | 28 | 72 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r +WT+eE++++++a k++G g W+ I +++g ++t+ q++ + k+
XP_015902737.1 28 REKWTEEEHQKFLEALKLYGHG-WRQIEEHVG-TKTAVQIRCHAXKF 72
789*******************.*********.********998776 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 2.02E-14 | 22 | 75 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 17.696 | 23 | 77 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.2E-10 | 24 | 71 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 4.3E-13 | 26 | 75 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.1E-10 | 27 | 75 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.5E-13 | 28 | 69 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 9.39E-11 | 30 | 73 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009651 | Biological Process | response to salt stress | ||||
| GO:0009723 | Biological Process | response to ethylene | ||||
| GO:0046686 | Biological Process | response to cadmium ion | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 211 aa Download sequence Send to blast |
MVLVIFDLIR MVFFSSQARK PYTIKKQREK WTEEEHQKFL EALKLYGHGW RQIEEHVGTK 60 TAVQIRCHAX KFFPKVVRGS TGSNEDSMKP IEIPLPRPKR KPMSPYPRKS VAFLKGMSVS 120 NRTERFSSPK EGTKSPISVL STLGSEAMDA PASDQHSKSS PSSTLCTTNM HSRSLSPLEK 180 ENDYTTSNSS AKEGKGSLPI LENVLCMINS N |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in phytochrome A-mediated cotyledon opening. Controlled by the central oscillator mediated by LHY and CCA1. Part of a regulatory circadian feedback loop. Regulates its own expression. {ECO:0000269|PubMed:14523250}. | |||||
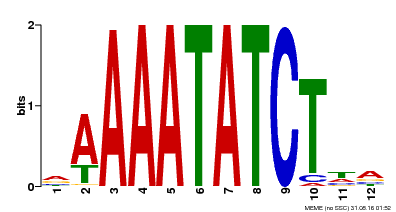
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00146 | DAP | Transfer from AT1G18330 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_015902737.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Circadian-regulation. Peak of transcript abundance near subjective dawn. Up-regulated transiently by red light and far red light. {ECO:0000269|PubMed:14523250, ECO:0000269|PubMed:19805390}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_015902737.2 | 1e-142 | LOW QUALITY PROTEIN: protein REVEILLE 7, partial | ||||
| Swissprot | B3H5A8 | 7e-44 | RVE7_ARATH; Protein REVEILLE 7 | ||||
| TrEMBL | A0A2P5CQF6 | 1e-71 | A0A2P5CQF6_TREOI; GAMYB transcription factor | ||||
| STRING | XP_010099677.1 | 1e-65 | (Morus notabilis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7313 | 32 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G18330.1 | 2e-46 | MYB_related family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



