 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_016435656.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 1242aa MW: 138799 Da PI: 9.2589 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 10.8 | 0.0016 | 1126 | 1151 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH.T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt.H 23
y C C++sFs+k +L H ++ +
XP_016435656.1 1126 YHCDleGCSMSFSTKQELSLHKKNvC 1151
889999***************99866 PP
| |||||||
| 2 | zf-C2H2 | 11 | 0.0013 | 1150 | 1173 | 2 | 23 |
EET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 2 kCp..dCgksFsrksnLkrHirtH 23
+Cp C+k F ++ +L++H r+H
XP_016435656.1 1150 VCPveGCKKKFFSHKYLVQHRRVH 1173
69999*****************99 PP
| |||||||
| 3 | zf-C2H2 | 12.2 | 0.00055 | 1209 | 1235 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHH..T CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirt..H 23
y+C+ Cg++F+ s++ rH r+ H
XP_016435656.1 1209 YVCTetGCGQTFRFVSDFSRHKRKtgH 1235
99********************99666 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00545 | 3.6E-16 | 19 | 60 | IPR003349 | JmjN domain |
| PROSITE profile | PS51183 | 14.991 | 20 | 61 | IPR003349 | JmjN domain |
| Pfam | PF02375 | 2.1E-15 | 21 | 54 | IPR003349 | JmjN domain |
| PROSITE profile | PS51184 | 31.698 | 180 | 349 | IPR003347 | JmjC domain |
| SMART | SM00558 | 1.8E-49 | 180 | 349 | IPR003347 | JmjC domain |
| SuperFamily | SSF51197 | 1.07E-24 | 194 | 338 | No hit | No description |
| Pfam | PF02373 | 3.0E-37 | 213 | 332 | IPR003347 | JmjC domain |
| SMART | SM00355 | 11 | 1126 | 1148 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 6.6E-5 | 1147 | 1177 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 11.572 | 1149 | 1178 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.012 | 1149 | 1173 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1151 | 1173 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 3.41E-9 | 1165 | 1205 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 2.8E-8 | 1178 | 1203 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 0.0014 | 1179 | 1203 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 10.741 | 1179 | 1208 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 1181 | 1203 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 5.76E-8 | 1197 | 1231 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 3.7E-9 | 1204 | 1232 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 10.907 | 1209 | 1240 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.19 | 1209 | 1235 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 1211 | 1235 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009741 | Biological Process | response to brassinosteroid | ||||
| GO:0009826 | Biological Process | unidimensional cell growth | ||||
| GO:0010228 | Biological Process | vegetative to reproductive phase transition of meristem | ||||
| GO:0033169 | Biological Process | histone H3-K9 demethylation | ||||
| GO:0035067 | Biological Process | negative regulation of histone acetylation | ||||
| GO:0048366 | Biological Process | leaf development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003676 | Molecular Function | nucleic acid binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1242 aa Download sequence Send to blast |
MAEASGNIEV FPWLKTLPVA PEYHPTLEEF QDPIAYIFKI EKEASKYGIC KIVPPVPAPP 60 KKTALAYLNR SLSARSGSNG GPTFTTRQQQ IGFCPRKHRP VKKPVWQSGE NYTVQEFQAK 120 AKAFEKNYLK KNFKKALTSP LEIETLFWKA TVDKPFSVEY ANDMPGSAFA PKKVGNVVTT 180 LADTEWNMRG VSRAKGSLLK FMKEEIPGVT SPMVYLAMMF SWFAWHVEDH DLHSLNYLHM 240 GAGKTWYGVP RDAAVAFEEV IRVHGYAGET NPLVTFATLG EKTTVMSPEV LLSAGIPCCR 300 LVQNAGDFVI TFPRAYHSGF SHGFNCGEAS NIATPGWLIV AKDAAIRRAS INCPPMVSHF 360 QLLYDLALSL CSRVPKNIRI EPRSSRLKDK KKSEGDMLVK ELFVEDLNCN NYLLHILGEG 420 SPVVLLPRHY SGISIGSNSV AGSQLKVNSR FPSLSSPDHE VKSKTDSASD ALMLGRKQRM 480 KQLASVSLEK GKHSSWHTGN RLPESGRDEA ESSPETEREN LDPERGMTYR CDTLSEHGLF 540 SCVTCGILCY TCVAIIQPTE AAAHHLMSSD YRNFNDWTGN VGGVTANSRD ANAAESDSSS 600 GWLVKRAPGG LINVPIESSD RIRKLNNESV GVLSSTKARK ETSSLGLLAL NYANSSDSDE 660 DEVEANIPVE ACESRQMDFE DEVSLRVIDP YANHRQRRAV SGGRNCQSLD NSVHLESGNL 720 PSGESNTLPD RSRHQLRSHQ VGANCIPFAH RGEIANSDGV APFDNGPMQF TSTSDEDSFR 780 IHVFCLQHAV QVEEQLRQVG GVHISLLCHP ADYPKLEAQA KKVAEELGRD HFWREISFRE 840 ATKEDEEMIQ SALEVEEAIH GNGDWTVKLD INLFYSANLS RSPLYSKQMP CNFIIYNAFG 900 RSSPDEKSEY TGTGRGSGKQ RRAVVAGKWC GKVWMSSQVH PLLAERRDTD EEQQQNNNSI 960 SARVKPEVKS ERPCEMTPTV KIVARTGKKR KSRVENKRNS KLLIADDSSV SNVPQQQRKT 1020 NLRSKRIKYE TPEPKEEDVD KKKRFGSPIN DDPDGGPSTR LRKRMPKPSK ESLVKPKPAP 1080 IKQQNESKKA EKGSKVKIPS SNSNSKKDPV MKANTSKKMK DKEGEYHCDL EGCSMSFSTK 1140 QELSLHKKNV CPVEGCKKKF FSHKYLVQHR RVHMDDRPLK CPWKGCKMTF KWAWARTEHI 1200 RVHTGARPYV CTETGCGQTF RFVSDFSRHK RKTGHISKKG RG |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6ip0_A | 4e-72 | 13 | 382 | 8 | 366 | Transcription factor jumonji (Jmj) family protein |
| 6ip4_A | 4e-72 | 13 | 382 | 8 | 366 | Arabidopsis JMJ13 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Histone demethylase that demethylates 'Lys-27' (H3K27me) of histone H3. Demethylates both tri- (H3K27me3) and di-methylated (H3K27me2) H3K27me (PubMed:21642989, PubMed:27111035). Demethylates also H3K4me3/2 and H3K36me3/2 in an in vitro assay (PubMed:20711170). Involved in the transcriptional regulation of hundreds of genes regulating developmental patterning and responses to various stimuli (PubMed:18467490). Binds DNA via its four zinc fingers in a sequence-specific manner, 5'-CTCTGYTY-3', to promote the demethylation of H3K27me3 and the regulation of organ boundary formation (PubMed:27111034, PubMed:27111035). Involved in the regulation of flowering time by repressing FLOWERING LOCUS C (FLC) expression (PubMed:15377760). Interacts with the NF-Y complexe to regulate SOC1 (PubMed:25105952). Mediates the recruitment of BRM to its target loci (PubMed:27111034). {ECO:0000269|PubMed:15377760, ECO:0000269|PubMed:18467490, ECO:0000269|PubMed:20711170, ECO:0000269|PubMed:21642989, ECO:0000269|PubMed:25105952, ECO:0000269|PubMed:27111034, ECO:0000269|PubMed:27111035}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00608 | ChIP-seq | Transfer from AT3G48430 | Download |
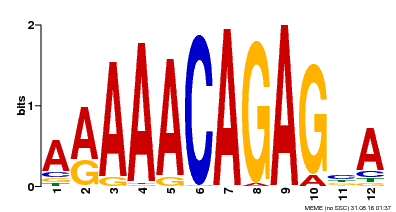
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975442 | 0.0 | HG975442.1 Solanum pennellii chromosome ch03, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_009613168.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6 isoform X1 | ||||
| Refseq | XP_016435656.1 | 0.0 | PREDICTED: lysine-specific demethylase REF6-like isoform X1 | ||||
| Swissprot | Q9STM3 | 0.0 | REF6_ARATH; Lysine-specific demethylase REF6 | ||||
| TrEMBL | A0A1S3X713 | 0.0 | A0A1S3X713_TOBAC; lysine-specific demethylase REF6-like isoform X1 | ||||
| STRING | XP_009613168.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Asterids | OGEA4988 | 23 | 31 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48430.1 | 0.0 | relative of early flowering 6 | ||||



