 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_016473141.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 697aa MW: 73938.7 Da PI: 6.2722 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 13.4 | 0.00015 | 447 | 467 | 35 | 55 |
TS-STCHHHHHHHHHHHHHHH CS
HLH 35 skKlsKaeiLekAveYIksLq 55
s++ +Ka++L +A+eY+k Lq
XP_016473141.1 447 SERADKASMLDEAIEYLKTLQ 467
7899****************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 3.79E-6 | 445 | 487 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 3.0E-5 | 446 | 475 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 8.30E-4 | 447 | 471 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 697 aa Download sequence Send to blast |
MPLSEFLKMA RGKLESGQQK ISSTTHLSSF PENELVELVW ENGQIMMQGQ SSSAKKNPIP 60 NKLPSNASKI GKFGVMDPML NDMPLSVPIG ELDLIQEDEG VPWLGYAADD SLQQDYCSQF 120 LPEISGLTTN EPAGQSEFGL INKRGSSDKM IRDSQSVPIH NAVNFDQRKV SPSSKFSLLC 180 SLAPQKGHAS IPSLGPGVSD IFSSKTSNNS VSVLGDSNQS QVSAGDVKSV KIQKQNMPGS 240 CSSLLNFSHF SRPATFVKAT KLQNITGVLA SDSSALEAKG CKEGENVTNS HNHVNAAAVE 300 DYLSFKKESG LQYPTSAVPS QRESRASGAS FHDRSSRVEQ SDNVFRESNS DNTPGQFTGA 360 KATTDTADGE GNAEHGVVCS SVCSGSSAER GSSDQPHNLK RKSRDNEDSG CPSEDVEEES 420 VGVKKSCPTR GGTGSKRSRA AEVHNLSERA DKASMLDEAI EYLKTLQLQV QIMSMGAGFC 480 VPPMMFPAGV QHMHAAQMPH FSPMGLGMGM GMGMGYGMGM LEMNGRSSGC PIFPIPSVQG 540 SHFPSPPIPA SAAYPGIAVS NRHAFAHPGQ GLPMPIPRPS LAPLAGQAST GAAVPLNVAR 600 VGVSVDRPSV PPILDSKNPV HKKNSQIVNN AECSLPINQT YSQVQATNEV LDKSALVQKN 660 DQLLDGTDSA ANILTNQTDV PGSEAGEFVR KDTATFS |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
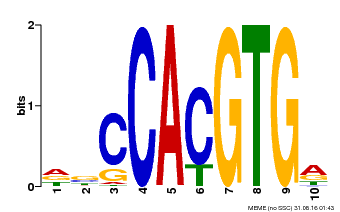
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016473141.1 | 0.0 | PREDICTED: transcription factor PIF3-like isoform X3 | ||||
| TrEMBL | A0A1S4A984 | 0.0 | A0A1S4A984_TOBAC; transcription factor PIF3-like isoform X3 | ||||
| STRING | XP_009774962.1 | 0.0 | (Nicotiana sylvestris) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 6e-28 | phytochrome interacting factor 3 | ||||



