 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_016474509.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; asterids; lamiids; Solanales; Solanaceae; Nicotianoideae; Nicotianeae; Nicotiana
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 997aa MW: 110829 Da PI: 6.0977 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.9 | 9.2e-41 | 142 | 219 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C +dls+ak+yhrrhkvCe+hska+ +lv +++qrfCqqCsrfh+l+efDe+krsCrrrLa+hn+rrrk+q+
XP_016474509.1 142 VCQVDDCGTDLSKAKDYHRRHKVCEMHSKASRALVGNVMQRFCQQCSRFHALQEFDEGKRSCRRRLAGHNKRRRKTQS 219
6**************************************************************************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 7.2E-33 | 137 | 204 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.086 | 140 | 217 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 7.72E-38 | 141 | 222 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 7.8E-30 | 143 | 216 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF48403 | 3.32E-7 | 770 | 883 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.8E-5 | 772 | 884 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.82E-6 | 773 | 883 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 997 aa Download sequence Send to blast |
MEASVGERYY HMGGPDLRGL GKRSLEWDVT DWKWDGDLFI ATPLRQNPSN NYQSRQFFPV 60 ETGNLVASSN SSSSCSDETN HGIEQQRREL EKRRRVIVVD ENESGGTLSL KLGGQVEPVA 120 EKRTKLAAAQ PVTSTPTTTR AVCQVDDCGT DLSKAKDYHR RHKVCEMHSK ASRALVGNVM 180 QRFCQQCSRF HALQEFDEGK RSCRRRLAGH NKRRRKTQSE SVVNNNSSND GQASGYSLMS 240 LLKMLSNMHS NGTNHTEDQD LLAHLLRSIA GQGSLNGDKS LSGLLQESSS MLNNRSILRN 300 PELASLISNG SQVPPRAKEH QFTNSAAEIP QKRLDAHDVR LEDARTASSQ SPGILFPPTQ 360 SNSQAYAPGR GSTTGRSKLI DFDLNDVYVD SDDNVEDIDR SPGQCPSWLQ QDSHQSSPPQ 420 TSGNSDSASA QSPSSSSGDT QTRTDRIVFK LFGKDPSDFP FVVRAQILDW LSHSPTEIES 480 YIRPGCVVLT VYLRLPESAW EELCYDLNSS LSRLLDVHGD DSFWTKGWIY ISVQDQIAFV 540 CDGQVLLDMS LPSGSDEHST ILSVRPIAVP VSGRAQFLVK GYNLSKPSTR LLCALESNYL 600 VPEANNEVEE HVDGIDKDDN LQSLNFTCSV PAVSGRGFIE VEDHGLSNSF FPFIVAEEDV 660 CSEIRMLESE LDLTSANYVK GQINNMEARN QAMDFIHELG WLLHRNNLKA RLEHFGPDAV 720 LYPLKRFKWL IDFCVDHEWC AVVKKLLNVL LDGTVGAGDS SFLKFALTEM GLLHRAVRRN 780 SRPLVELLLT YTPEKVADEL SSEYQSLVGV DGEFLFRPDC VGPAGLTPLH VAAGIDGSED 840 VLDALTDDPG KVAIEAWKNT RDSTGFTPED YARLRGHYSY IHLVQRKISK KATSGHIVVD 900 IPIVPSVENS NQKEEFATTS LEISMTERRP ISRPCGLCHK KLAYGSRSRS LLYRPAMFSM 960 VAMAAVCVCV ALLFRGSPEV LYIFRPFRWE MVDFGTS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 6e-31 | 134 | 216 | 2 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 91 | 95 | KRRRV |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
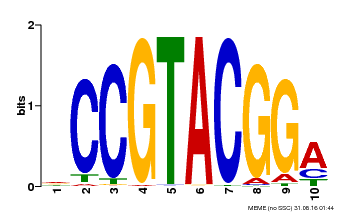
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' of AP1 promoter. Binds specifically to the 5'-GTAC-3' core sequence. {ECO:0000269|PubMed:10524240, ECO:0000269|PubMed:16095614}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00603 | PBM | Transfer from AT2G47070 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | HG975440 | 0.0 | HG975440.1 Solanum pennellii chromosome ch01, complete genome. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016474509.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 1 isoform X2 | ||||
| Swissprot | Q9SMX9 | 0.0 | SPL1_ARATH; Squamosa promoter-binding-like protein 1 | ||||
| TrEMBL | A0A1S4AD26 | 0.0 | A0A1S4AD26_TOBAC; squamosa promoter-binding-like protein 1 isoform X2 | ||||
| STRING | XP_009592086.1 | 0.0 | (Nicotiana tomentosiformis) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47070.1 | 0.0 | squamosa promoter binding protein-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



