 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_016647596.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 874aa MW: 98934.1 Da PI: 7.6789 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 163.1 | 4.9e-51 | 3 | 118 | 3 | 118 |
CG-1 3 kekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrr 97
+ k+rwl+++ei+aiL n++ +++ ++ + p+sg+++L++rk++r+frkDG++wkkk dgktv+E+he+LKvg+ e +++yYah+e+ ptf rr
XP_016647596.1 3 EAKSRWLRPNEIHAILYNHKYFTIYVKPVNLPQSGTIVLFDRKMLRNFRKDGHNWKKKNDGKTVKEAHEHLKVGNEERIHVYYAHGEDSPTFVRR 97
459******************************************************************************************** PP
CG-1 98 cywlLeeelekivlvhylevk 118
cywlL+++le+ivlvhy+e++
XP_016647596.1 98 CYWLLDKSLEHIVLVHYRETQ 118
******************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 75.892 | 1 | 123 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 2.4E-75 | 1 | 118 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.4E-45 | 3 | 116 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 1.26E-13 | 328 | 415 | IPR014756 | Immunoglobulin E-set |
| CDD | cd00204 | 4.77E-14 | 488 | 622 | No hit | No description |
| Gene3D | G3DSA:1.25.40.20 | 1.6E-16 | 512 | 626 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 6.22E-17 | 525 | 625 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 5.5E-7 | 525 | 592 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 15.3 | 530 | 634 | IPR020683 | Ankyrin repeat-containing domain |
| SMART | SM00248 | 9.2E-6 | 563 | 592 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 11.621 | 563 | 595 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 1600 | 602 | 632 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 1.42E-5 | 702 | 777 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| PROSITE profile | PS50096 | 7.73 | 711 | 737 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 26 | 726 | 748 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.053 | 727 | 756 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.022 | 749 | 771 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.164 | 750 | 774 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 7.8E-4 | 751 | 771 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 46 | 829 | 851 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.199 | 830 | 859 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 874 aa Download sequence Send to blast |
MEEAKSRWLR PNEIHAILYN HKYFTIYVKP VNLPQSGTIV LFDRKMLRNF RKDGHNWKKK 60 NDGKTVKEAH EHLKVGNEER IHVYYAHGED SPTFVRRCYW LLDKSLEHIV LVHYRETQEL 120 QGSPVTPVNS NNSSSVSDPS APWLLSEELD SGANKSYCAG ENELSEPGDG LTVKNHEKRL 180 HDINTLEWEE LLITNDSKGD IVSCYDQQNQ VVGNGFISGG ASVISAEMSA FDNLTNPTSR 240 SDNVQFNLLD SPYVPTVEKT TYDSLDVLVN DGLHSQDSFG RWINQVMADP PGSVEDPALE 300 SSSLAAQNSF ASPSADHLQS SVPHQIFNIT DLSPAWAFSN EKTKILITGF FHQEYLHLAK 360 SDLLCICGDV CLRAEIVQAG VYRCFVPPHL PRVVNLFMSI DGHKPISLVL NFEYRAPVLS 420 DPIISSEENK WEEFQAQMRL AYLLFSSSKN LNIVSNKVLP NALKEAKKFS HRTSHISNSW 480 AYLMKAVEDN KTPLPLAKDG LFELILKNRL KDWLLEKVVA SSTTKEYDAY GQGVIHLCAI 540 LEYTWAVRLF SWSGLSLDFR DRRGWTALHW AAYCGREKMV AVLLSAGAKP NLVTDPSSEN 600 PGGCTAADLA AMKGYDGLAA YLSEKALVEQ FKDMSMAGNA SGSLQTSSNY AGNSENLSED 660 EIHLKDTLAA YRTAADAAAR IQAAFRENSL KLKAKAVQYS TPEAEARGII AALKIQHAFR 720 NYDTRKKIKA AARIQYRFRT WKMRQEFLSL RRQAIKIQAA FRGFQVRRQY RKVLWSVGVL 780 EKAVLRWRFK RRGLRGLNVA PVEVDVDQKQ ESDTEEDFYR ASRKQAEERI ERSVVRVQAM 840 FRSKKAQEEY SRMKLTHIEA KLEFEELLNP DLDS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator (PubMed:14581622). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in response to cold. Contributes together with CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:28351986). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:28351986, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
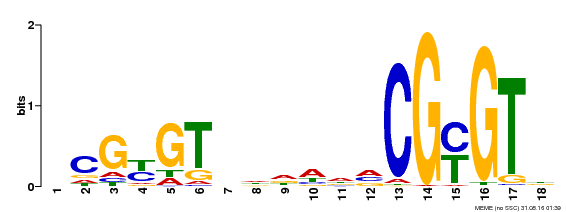
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_016647596.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By heat shock, UVB, wounding, abscisic acid, H(2)O(2) and salicylic acid. {ECO:0000269|PubMed:12218065}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC208139 | 1e-38 | AC208139.1 Populus trichocarpa clone JGIACSB13-D20, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008223308.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 | ||||
| Refseq | XP_016647596.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 5-like isoform X3 | ||||
| Swissprot | O23463 | 0.0 | CMTA5_ARATH; Calmodulin-binding transcription activator 5 | ||||
| TrEMBL | A0A251QWG4 | 0.0 | A0A251QWG4_PRUPE; Uncharacterized protein | ||||
| STRING | XP_008223308.1 | 0.0 | (Prunus mume) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 0.0 | calmodulin binding;transcription regulators | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



