 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_016648488.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | NAC | ||||||||
| Protein Properties | Length: 371aa MW: 42052 Da PI: 7.8059 | ||||||||
| Description | NAC family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | NAM | 173.7 | 5.5e-54 | 11 | 141 | 1 | 129 |
NAM 1 lppGfrFhPtdeelvveyLkkkvegkkleleevikevdiykvePwdLpk..kvka.eekewyfFskrdkkyatgkrknratksgyWkatgkdkev 92
+ppGfrFhPtdeel+ +yL+kkv+ + ++l +vi+evd++k+ePwdL++ ++ + ++ewyfFs++dkky+tg+r+nrat++g+Wkatg+dk++
XP_016648488.1 11 VPPGFRFHPTDEELLYYYLRKKVSYEAIDL-DVIREVDLNKLEPWDLKEkcRIGSgPQNEWYFFSHKDKKYPTGTRTNRATTAGFWKATGRDKAI 104
69****************************.9***************952444443456************************************ PP
NAM 93 lskkgelvglkktLvfykgrapkgektdWvmheyrle 129
+ ++++ +g++ktLvfy+grap+g+ktdW+mheyrle
XP_016648488.1 105 HLANSKRIGMRKTLVFYTGRAPHGQKTDWIMHEYRLE 141
***********************************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101941 | 6.54E-58 | 5 | 171 | IPR003441 | NAC domain |
| PROSITE profile | PS51005 | 56.488 | 11 | 171 | IPR003441 | NAC domain |
| Pfam | PF02365 | 5.1E-28 | 12 | 140 | IPR003441 | NAC domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003002 | Biological Process | regionalization | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009834 | Biological Process | plant-type secondary cell wall biogenesis | ||||
| GO:0010455 | Biological Process | positive regulation of cell fate commitment | ||||
| GO:0048829 | Biological Process | root cap development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 371 aa Download sequence Send to blast |
MMASGNGQLT VPPGFRFHPT DEELLYYYLR KKVSYEAIDL DVIREVDLNK LEPWDLKEKC 60 RIGSGPQNEW YFFSHKDKKY PTGTRTNRAT TAGFWKATGR DKAIHLANSK RIGMRKTLVF 120 YTGRAPHGQK TDWIMHEYRL EDDNISADQV QVVGLLVXXX XGWVVCRVFK KKNHNRGYQP 180 EFAHHQDQEH DHFTNMKASG SHAPVLLEHH EPKQNHHHLE ALYDYTSNFD HAGSMHLPQL 240 FSPESAVASS FVSPNTINLN TMDIECSQNL LRLTSTAAAA AAAAAGCGQL MQQQERSLIN 300 GGDWSFLDKL LASHQSLDHH HSSHTKCNNP SSSAQHQLNN SHEHVGTSST TQQRFPFQYL 360 GCGTDLLKFS K |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 3ulx_A | 1e-49 | 7 | 174 | 11 | 171 | Stress-induced transcription factor NAC1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription regulator. Together with BRN1 and BRN2, regulates cellular maturation of root cap. Represses stem cell-like divisions in the root cap daughter cells, and thus promotes daughter cell fate. Inhibits expression of its positive regulator FEZ in a feedback loop for controlled switches in cell division plane. Promotes the expression of genes involved in secondary cell walls (SCW) biosynthesis. {ECO:0000269|PubMed:19081078, ECO:0000269|PubMed:20197506}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00250 | DAP | Transfer from AT1G79580 | Download |
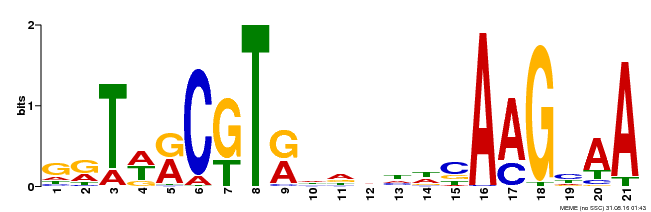
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_016648488.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By FEZ in oriented-divised root cap stem cells. {ECO:0000269|PubMed:19081078}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016648488.1 | 0.0 | PREDICTED: protein SOMBRERO | ||||
| Swissprot | Q9MA17 | 1e-124 | SMB_ARATH; Protein SOMBRERO | ||||
| TrEMBL | A0A314ZIC4 | 0.0 | A0A314ZIC4_PRUYE; Protein SOMBRERO isoform X1 | ||||
| STRING | XP_008225961.1 | 0.0 | (Prunus mume) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7204 | 34 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G79580.3 | 1e-112 | NAC family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



