 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | XP_016651430.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Rosales; Rosaceae; Maloideae; Amygdaleae; Prunus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 680aa MW: 76691 Da PI: 6.2597 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 143.3 | 2.2e-44 | 68 | 199 | 2 | 134 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslqs 96
+g++++++E+E++k+RER RR i+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGwvve+DGttyr++ p + a+g + sp ss
XP_016651430.1 68 NKGKREREREKERTKLRERLRRSITSRMLAGLRQYGNFPLPARADMNDVLAALAREAGWVVEADGTTYRQSPPPS--QIAMGMRSVESPVSSGVR 160
5799*****************************************************************765555..778888888999999886 PP
DUF822 97 slkssalaspvesysaspksssfpspssldsislasa..a 134
++ks + + p ++ s++ + ++++sp+slds+ +a+ +
XP_016651430.1 161 ATKSLECHPP-HNPSSVVRIDESLSPASLDSVVIAERdtT 199
6666666655.55677889999**********99987543 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 2.0E-45 | 69 | 203 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Gene3D | G3DSA:3.20.20.80 | 6.2E-169 | 241 | 672 | IPR013781 | Glycoside hydrolase, catalytic domain |
| SuperFamily | SSF51445 | 1.12E-156 | 242 | 672 | IPR017853 | Glycoside hydrolase superfamily |
| Pfam | PF01373 | 1.0E-81 | 265 | 636 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.4E-54 | 278 | 292 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.4E-54 | 299 | 317 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.4E-54 | 321 | 342 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.4E-54 | 414 | 436 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.4E-54 | 487 | 506 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.4E-54 | 521 | 537 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.4E-54 | 538 | 549 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.4E-54 | 556 | 579 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 2.4E-54 | 594 | 616 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 680 aa Download sequence Send to blast |
MNDDQSLRQD LDPHSDHSSD YLPHHPHHPS HPLQPQQQPQ PQPRRPRGFA ATAAGSVGPT 60 SPSTPTTNKG KREREREKER TKLRERLRRS ITSRMLAGLR QYGNFPLPAR ADMNDVLAAL 120 AREAGWVVEA DGTTYRQSPP PSQIAMGMRS VESPVSSGVR ATKSLECHPP HNPSSVVRID 180 ESLSPASLDS VVIAERDTTK NDKFSSGGSP INSVEADQLM QDLPIRPGNH GIGTTAFSTP 240 DPYVPLYVTL ATGFINNYCQ LVDPEGLRQE LTHMQSLSVD GVVVDCWWGI VEGWSPQKYV 300 WSGYRGLFNI LREFKFKFQV VMAFHEYGRS ESGEALIPLP QWILEIGKEN QDIFFTDREG 360 RRNTECLSWG IDKERVLHGR TGVEVYFDFM RSFRTEFDDL FAEGLISAVE IGLGSSGELK 420 YPSFSGRMGW RYPGVGEFQC YDRYLQQSLR RAAKLRGHSF WARGPDNAGE YNSRPHETGF 480 FCERGDYDSY YGRFFLHWYA QSLIDHADNV LSLASLAFDE TKIIVKVPAV YWWYKTSSHA 540 AELTSGYYNP TNQNGYSPVF EVLKKHSVTV KFVCSGLHMS SQDNDEALAD PEGLSWQVLN 600 LAWDQGLLVA GENALSCYDR EGCMRIVEMV KPRNDPDHRH FSFFVYQQPA PLVQGAICFS 660 ELDFFIKCMH GEIAGDLVSC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-120 | 243 | 633 | 11 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
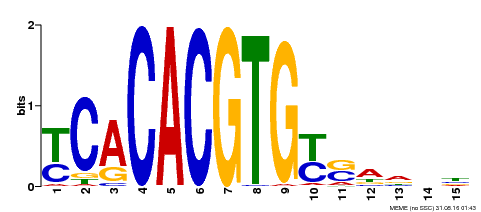
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | XP_016651430.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016651430.1 | 0.0 | PREDICTED: beta-amylase 8 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A251P205 | 0.0 | A0A251P205_PRUPE; Beta-amylase | ||||
| STRING | EMJ12027 | 0.0 | (Prunus persica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11902 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.1 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



