 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00001.1.g09690.1.sm.mkhc | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 429aa MW: 48685.5 Da PI: 10.4246 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 153.7 | 4.1e-48 | 32 | 136 | 3 | 106 |
CG-1 3 ke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvev 80
e +rw +++ei+a+L+n++k+++ ++ ++p sg+++Ly+rk+vr+frkDG++wkkkkdgktv+E+hekLK+g+ e
Zjn_sc00001.1.g09690.1.sm.mkhc 32 AEaASRWFRPNEIYAVLANYTKFQIIPQPIDKPASGTVVLYDRKVVRNFRKDGHNWKKKKDGKTVQEAHEKLKIGNEEK 110
44499************************************************************************** PP
CG-1 81 lycyYahseenptfqrrcywlLeeel 106
+++yYa++e++p+f rrcywlL++e+
Zjn_sc00001.1.g09690.1.sm.mkhc 111 VHVYYARGEDDPNFFRRCYWLLDKEK 136
***********************993 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 63.441 | 27 | 154 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.0E-50 | 30 | 139 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 4.8E-40 | 34 | 137 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:1.25.40.20 | 2.4E-4 | 134 | 221 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF52540 | 2.15E-7 | 237 | 338 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.85 | 287 | 309 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.766 | 289 | 317 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.003 | 290 | 308 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0089 | 310 | 332 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 10.31 | 311 | 335 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 3.0E-4 | 312 | 332 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 5.6 | 391 | 413 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.157 | 393 | 421 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.03 | 394 | 413 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 429 aa Download sequence Send to blast |
MAGGAGGQDP LVASEIHGFL TCADLNYEKL MAEAASRWFR PNEIYAVLAN YTKFQIIPQP 60 IDKPASGTVV LYDRKVVRNF RKDGHNWKKK KDGKTVQEAH EKLKIGNEEK VHVYYARGED 120 DPNFFRRCYW LLDKEKMVAA LLSAGANPSL VTDPTPDAPG GYTAADLAAA RGYDGLAAYL 180 AEKGLTAHFE AMSLSKDTGR SPSRRNSIKQ HTKEFETLSE QELCLRESLA AYRNAADAAS 240 NIQAALRERT LKLQTKAIQL ANPENEAAAI VAAMRIQHAF RNYNRKKVMR AAARIQSHFR 300 TWKIRRDFMN IRRQAIKIQA AYRGHQVRRQ YRKVIWSVGV VEKAILRWRK KRKGLRGIAT 360 GMAVAMATDA EPASTAEEDY YQIGRQQAED RFNRSVVRVQ ALFRSYRAQQ EYRRMKVAHE 420 EAKVEFSGN |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro. Binds to the DNA consensus sequence 5'-T[AC]CG[CT]GT[GT][GT][GT][GT]T[GT]CG-3'. {ECO:0000269|PubMed:16192280}. | |||||
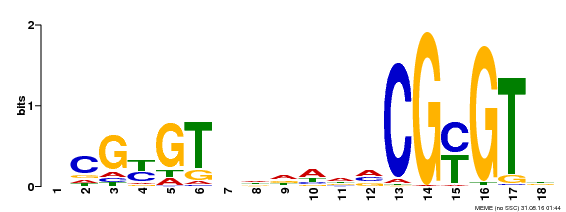
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00435 | DAP | Transfer from AT4G16150 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004957727.1 | 1e-171 | calmodulin-binding transcription activator 6 | ||||
| Swissprot | Q7XHR2 | 1e-163 | CBT_ORYSJ; Calmodulin-binding transcription activator CBT | ||||
| TrEMBL | C4JAR3 | 1e-175 | C4JAR3_MAIZE; Uncharacterized protein | ||||
| STRING | Si028862m | 1e-171 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9831 | 29 | 33 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16150.1 | 1e-81 | calmodulin binding;transcription regulators | ||||



