 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00005.1.g03000.1.sm.mkhc | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 633aa MW: 67626.9 Da PI: 6.6549 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.4 | 8e-16 | 385 | 431 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K +Ka++L +A+eY+k Lq
Zjn_sc00005.1.g03000.1.sm.mkhc 385 VHNLSERRRRDRINEKMRALQELIPNC-----NKIDKASMLDEAIEYLKTLQ 431
5*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.80E-17 | 378 | 435 | No hit | No description |
| SuperFamily | SSF47459 | 1.96E-20 | 379 | 440 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.981 | 381 | 430 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.7E-13 | 385 | 431 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 2.3E-20 | 385 | 439 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 2.1E-17 | 387 | 436 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 633 aa Download sequence Send to blast |
MHACAPPVHG CRSDGNDFAE LLWENGQAVV HGRRKQPLTA FPPFTCGGAS SSKAQEKHPG 60 SDPVALLKTT GVYGAGTMAP SAHDLYSGLD TSRGNGDLDD TVPWIHYPII EDDDEGAAPA 120 LADSYSPDFF SELQAAAAVA VNLTPPPPSV QHTADNRSAG AATMAGEPEP SKESRRISVS 180 APRPEPQAEF KANKQPRLGG GGDALMNFSL FSRPAAMARA SLQQSAQRPP PPLGTDKPPG 240 VTVTSSTRVE STVVQSSSGP RTAPVHTDQR MAWPQAKEVR FACTAAPTSM AAGIMQQELP 300 RDRIANDMAP QRRVEAKKVP EAAIAASSVC SGNGAGIGND ESWRQQKRKS QAECSASQDD 360 DLEDESGGLR RSGSRSTKRS RTAEVHNLSE RRRRDRINEK MRALQELIPN CNKIDKASML 420 DEAIEYLKTL QLQVQMMSMG SGLCIPPMLL PPTMQHLQIP TMAHFPHLGM GLGYGMGIFD 480 MNSTTTVPLP PMPGAHFPCQ MIPGTAPHAI GIPGRNTLPM FGVPGQPIHP LVPGVQPFPS 540 LASLPVRPNL APQVSATVAY MVQEQQQVSN QQQQSLNDEG TQGASTRDPQ IQTVLQVDNQ 600 HFSVPSSTQS ESSHFLDSGG DKTNTAVRNG VET |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 389 | 394 | ERRRRD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that may act as negative regulator of phyB-dependent light signal transduction. {ECO:0000269|PubMed:17485859}. | |||||
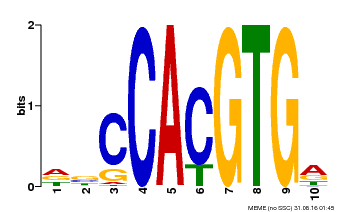
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by light in dark-grown etiolated seedlings. {ECO:0000269|PubMed:17485859}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004967640.1 | 0.0 | transcription factor APG isoform X1 | ||||
| Refseq | XP_004967641.1 | 0.0 | transcription factor APG isoform X1 | ||||
| Refseq | XP_004967642.1 | 0.0 | transcription factor APG isoform X1 | ||||
| Refseq | XP_004967643.1 | 0.0 | transcription factor APG isoform X1 | ||||
| Refseq | XP_004967644.1 | 0.0 | transcription factor APG isoform X1 | ||||
| Refseq | XP_022682497.1 | 0.0 | transcription factor APG isoform X1 | ||||
| Swissprot | Q0JNI9 | 0.0 | PIL15_ORYSJ; Transcription factor PHYTOCHROME INTERACTING FACTOR-LIKE 15 | ||||
| TrEMBL | A0A3L6R267 | 0.0 | A0A3L6R267_PANMI; Transcription factor APG-like isoform X1 | ||||
| STRING | Si000700m | 0.0 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3527 | 37 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 4e-42 | phytochrome interacting factor 3 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



