 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00007.1.g05310.1.sm.mk | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 339aa MW: 36933.2 Da PI: 9.8102 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 73.2 | 2.9e-23 | 58 | 123 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwkg 71
rk+ksL+ll+++f++l+++ + +++ l+++a++L v +rRRiYDi+NVLe+++++ +k+kn ++w+g
Zjn_sc00007.1.g05310.1.sm.mk 58 RKQKSLGLLCSNFVALYNRDHVEHIGLDKAAQSL---GV--ERRRIYDIVNVLESIGILVRKAKNLYTWIG 123
79*****************999************...**..****************************98 PP
| |||||||
| 2 | E2F_TDP | 76.4 | 2.8e-24 | 202 | 281 | 1 | 70 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvse.....dvknkrRRiYDilNVLealnliek.....kekneirwk 70
rkeksL+lltq+f+kl+ ++e ++++l+e++k L+ e ++++k+RR+YDi+NVL++lnl+ek ++k+++rw
Zjn_sc00007.1.g05310.1.sm.mk 202 RKEKSLGLLTQNFVKLFLTKEVDTISLDEASKLLLGEgheetNMRTKVRRLYDIANVLSSLNLLEKvhqanTKKPAFRWL 281
79*****************999************************************************999******6 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 7.1E-26 | 54 | 124 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 6.35E-16 | 57 | 122 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 3.0E-28 | 58 | 123 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 1.5E-20 | 59 | 123 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 3.4E-22 | 199 | 282 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 7.0E-31 | 202 | 282 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 1.9E-19 | 203 | 281 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| SuperFamily | SSF46785 | 1.63E-11 | 204 | 281 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032876 | Biological Process | negative regulation of DNA endoreduplication | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 339 aa Download sequence Send to blast |
MATTPAMAAA ASSVAQACAA AAAAAAGMES ALWQQERPRA KVAEGGGPKG ARHHAYSRKQ 60 KSLGLLCSNF VALYNRDHVE HIGLDKAAQS LGVERRRIYD IVNVLESIGI LVRKAKNLYT 120 WIGFDGIPIA LRELKDRALR EKSGLAPSQM EQLSAANVSD DEDDDKLGDT DGDTDSEKLS 180 QTVDNPSDKP GASQCRLRSD HRKEKSLGLL TQNFVKLFLT KEVDTISLDE ASKLLLGEGH 240 EETNMRTKVR RLYDIANVLS SLNLLEKVHQ ANTKKPAFRW LAKAGKPKTE TGVAFAVSPS 300 AKSRLVIFFP ITLKLGSLGI LIWRKAATLC GNTWGSQER |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4yo2_A | 1e-37 | 57 | 269 | 3 | 223 | Transcription factor E2F8 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Inhibitor of E2F-dependent activation of gene expression. Binds specifically the E2 recognition site without interacting with DP proteins and prevents transcription activation by E2F/DP heterodimers. Controls the timing of endocycle onset and inhibits endoreduplication. {ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:11867638, ECO:0000269|PubMed:15649366, ECO:0000269|PubMed:18787127}. | |||||
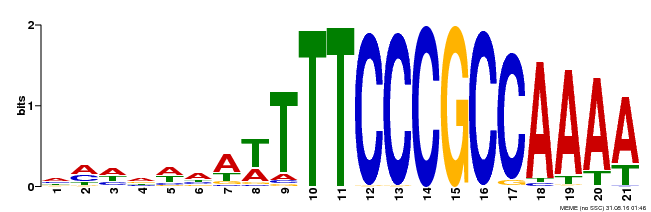
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00397 | DAP | Transfer from AT3G48160 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022682371.1 | 1e-154 | E2F transcription factor-like E2FE | ||||
| Swissprot | Q8LSZ4 | 4e-77 | E2FE_ARATH; E2F transcription factor-like E2FE | ||||
| TrEMBL | A0A368PRM9 | 1e-153 | A0A368PRM9_SETIT; Uncharacterized protein | ||||
| STRING | Si019635m | 1e-149 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3185 | 38 | 84 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48160.1 | 1e-78 | DP-E2F-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



