 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00008.1.g01280.1.am.mkhc | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 414aa MW: 44872.9 Da PI: 6.1964 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 91.6 | 6.8e-29 | 268 | 321 | 1 | 54 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYR 54
k rlrWt eLHerFveav++L G+ekAtPk +l+lmkv+gLt++hvkSHLQ R
Zjn_sc00008.1.g01280.1.am.mkhc 268 KSRLRWTLELHERFVEAVNKLDGPEKATPKGVLKLMKVEGLTIYHVKSHLQVQR 321
68*************************************************877 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.165 | 265 | 325 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.4E-26 | 266 | 321 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.97E-14 | 266 | 322 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.8E-22 | 268 | 322 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 8.8E-7 | 270 | 319 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 8.9E-9 | 316 | 344 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 414 aa Download sequence Send to blast |
MSSRSALAVE QITAPDKTVH ASTATPPLLH KLFDAKLDHQ VLVDDTLSST SQSSNIKTEL 60 MRSSSFSKSL TLNLQKGSPE PDPESPLSHV SHPNFSDPIP SNSSTFCTTL FSSSLANPER 120 SRRIGTLPFL PHPPKCEQQV SAGQSLSSSL LLSRDIGNSL DDTEHSNDLK VLLNLSGDGS 180 DGSYHEENNA LAFAEHMEFQ FLSEQLGIAI TDNEESPRLD DIYDTPPPQL SALPVSSCSN 240 QSIQNPGSPV KVQLNSSRSS SGSVATNKSR LRWTLELHER FVEAVNKLDG PEKATPKGVL 300 KLMKVEGLTI YHVKSHLQVQ RQLQLRIEEH AKYLQKLLEE QQKTGNTSPP KTLSKTQAES 360 PESVSKESGE SEAGNSSPPS SKKRVPDVDM ESNLLAGSKR TKVQVGSGSE GSCS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 9e-23 | 268 | 326 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 9e-23 | 268 | 326 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 9e-23 | 268 | 326 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 9e-23 | 268 | 326 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 9e-23 | 268 | 326 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 9e-23 | 268 | 326 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 9e-23 | 268 | 326 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 9e-23 | 268 | 326 | 2 | 60 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor involved in phosphate starvation signaling (PubMed:26082401). Binds to P1BS, an imperfect palindromic sequence 5'-GNATATNC-3', to promote the expression of inorganic phosphate (Pi) starvation-responsive genes (PubMed:26082401). Functionally redundant with PHR1 and PHR2 in regulating Pi starvation response and Pi homeostasis (PubMed:26082401). {ECO:0000269|PubMed:26082401}. | |||||
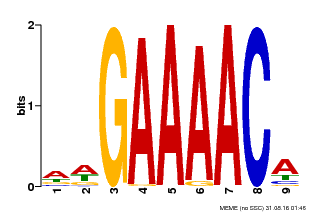
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00354 | DAP | Transfer from AT3G13040 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated under Pi starvation conditions. {ECO:0000269|PubMed:26082401}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025812859.1 | 0.0 | protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| Refseq | XP_025812860.1 | 0.0 | protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| Refseq | XP_025812862.1 | 0.0 | protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| Refseq | XP_025812863.1 | 0.0 | protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| Swissprot | Q6YXZ4 | 1e-127 | PHR3_ORYSJ; Protein PHOSPHATE STARVATION RESPONSE 3 | ||||
| TrEMBL | A0A3L6PFL7 | 0.0 | A0A3L6PFL7_PANMI; Uncharacterized protein | ||||
| STRING | Pavir.J27649.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4526 | 37 | 69 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G13040.2 | 5e-59 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



