 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00011.1.g01330.1.sm.mkhc | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 296aa MW: 32226.8 Da PI: 10.106 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 36.7 | 1e-11 | 39 | 90 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT.....TTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmg.....kgRtlkqcksrwqky 47
+++WT+eE++ l ++v ++G g W+tI + R+ ++k++w+++
Zjn_sc00011.1.g01330.1.sm.mkhc 39 KQKWTQEEEDALRRGVLKHGAGKWRTIQKDPEfsptlSSRSNIDLKDKWRNL 90
79***********************************9************96 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.905 | 34 | 94 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.66E-15 | 36 | 92 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 4.6E-8 | 38 | 93 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-14 | 39 | 90 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 1.6E-8 | 39 | 90 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 3.60E-19 | 40 | 90 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 296 aa Download sequence Send to blast |
MNTRRIGEDR PLLVYVECGW TLEKPQIAGG EARAMGAPKQ KWTQEEEDAL RRGVLKHGAG 60 KWRTIQKDPE FSPTLSSRSN IDLKDKWRNL SFSAGQGSRE KTRVPKITGP SSSALPAPQA 120 LLLPASNKIA EASAPADAEK KSQDGKASPK ESTAWRPHDA LCEDKIKTRC LQRGWKGLKV 180 DNKFYKLTDS FATKTPALVK ASTKKQKDPS KPLKASKNLG IFAAVSPALE AAAAAAAKVA 240 DAGAKAHLAH EHMMEADRIL KMAEETESLL TLAAEIYDRC SSGEIITVNP VSQKEF |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds preferentially double-stranded telomeric repeats, but may also bind to the single telomeric strand. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00652 | PBM | Transfer from LOC_Os01g51154 | Download |
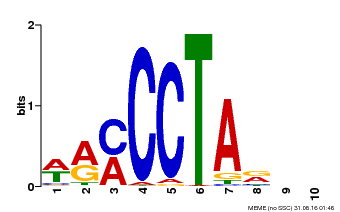
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | FP094524 | 2e-72 | FP094524.1 Phyllostachys edulis cDNA clone: bphylf024j07, full insert sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | NP_001105225.1 | 1e-100 | single myb histone 3 | ||||
| Swissprot | Q6WLH4 | 1e-101 | SMH3_MAIZE; Single myb histone 3 | ||||
| TrEMBL | A0A0A9PJS4 | 1e-109 | A0A0A9PJS4_ARUDO; Smh4 | ||||
| STRING | Pavir.Eb02802.1.p | 2e-96 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9247 | 34 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G49950.3 | 2e-29 | telomere repeat binding factor 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



