 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00012.1.g09180.1.sm.mkhc | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 825aa MW: 90981.4 Da PI: 5.8671 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 111.8 | 4.2e-35 | 187 | 262 | 1 | 76 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkk 76
+Cqv+gCead+ e+k yhrrh+vC +++a++vl++g+++r+CqqC++fh l +fDe+krsCrr+L++hn+rrr+k
Zjn_sc00012.1.g09180.1.sm.mkhc 187 RCQVPGCEADIRELKGYHRRHRVCLRCAHAAAVLLDGVQKRYCQQCGKFHVLLDFDEDKRSCRRKLERHNKRRRRK 262
6************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 5.4E-27 | 184 | 249 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 29.615 | 185 | 262 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.01E-33 | 186 | 266 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 8.8E-27 | 188 | 262 | IPR004333 | Transcription factor, SBP-box |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0055070 | Biological Process | copper ion homeostasis | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 825 aa Download sequence Send to blast |
MDAPYSGGGG VDAGEPVWDW GNLLDFVVQD DDSLVLPWDD VAGTGAAGPT EAGASLLATP 60 PPQPAEVEAN QEPEPEPEPE PVLPPLPLHV QGIGRRVRKR DPRLVCPNYL AGRVPCACPE 120 VDEMAAAAEV EDVATELLAG ARKKVKTAGR RGKAGAVGGA GGSGGGVSTT GGGSGGGGRA 180 VVAAEMRCQV PGCEADIREL KGYHRRHRVC LRCAHAAAVL LDGVQKRYCQ QCGKFHVLLD 240 FDEDKRSCRR KLERHNKRRR RKPDSKVPFE KEMDEHLDLS ADVSGDGELR EENIECTTSE 300 MLETVLSNKV LDRETPMGSE DVLSSPTCTQ PNLQNEQSKS IVTFAASVEA SSGAKQESVK 360 LNNSPIHDLK TAYSSSCPTG RISFKLYDWN PAEFPRRLRH QIFEWLASMP VELEGYIRPG 420 CTILTVFIAM PQHMWDKLSD DTANLLRNLV NCPNSLLLGK GNFFIHIDNM IFQVLKDGAT 480 LMSTRLEMQA PRIHYVHPTW FEAGKPVELF VCGSSLDQPK FRSLLSFDGN YLKHDCFRVA 540 SLDAFDCVEK DDLKLDSQHE MFRISISGSR PDTQGPAFVE VENMFGLSNF VPILFGSRQL 600 CSELAKIQDV LCGSCKNNDI FEEFPSASSD PCECKKVKSA AMSGFLIDIG WLIRKPSSEE 660 FKNVLSLTNI QRWVYMLKFL IQNDFLNVLE IIVKSMDNFI GSEILSNLEK GRLEDHVATF 720 LAYLNHAQSI VNQKAKHGEE TQLETRLVTD KFPKHQSLGT SVPLDKGKTT GLGGHNDLLL 780 TSAACEEEVT MPLMTKDATH RQCCHPDISA RWLKHSRVGP QEHDS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul5_A | 2e-36 | 187 | 266 | 5 | 84 | squamosa promoter binding protein-like 7 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 244 | 262 | KRSCRRKLERHNKRRRRKP |
| 2 | 248 | 260 | RRKLERHNKRRRR |
| 3 | 255 | 262 | NKRRRRKP |
| 4 | 256 | 261 | KRRRRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00604 | PBM | Transfer from AT5G18830 | Download |
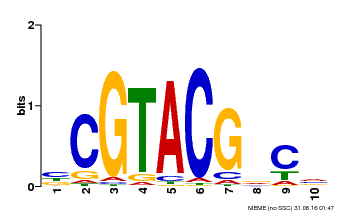
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025805456.1 | 0.0 | squamosa promoter-binding-like protein 9 | ||||
| Swissprot | Q6I576 | 0.0 | SPL9_ORYSJ; Squamosa promoter-binding-like protein 9 | ||||
| TrEMBL | A0A3L6T078 | 0.0 | A0A3L6T078_PANMI; Squamosa promoter-binding-like protein 9 | ||||
| STRING | GRMZM2G109354_P01 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP6922 | 33 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G18830.2 | 1e-136 | squamosa promoter binding protein-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



