 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00014.1.g02690.1.am.mk | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 708aa MW: 78112.6 Da PI: 7.7888 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 27.1 | 7e-09 | 447 | 485 | 17 | 55 |
HHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 17 elFeknrypsaeereeLAkklgLterqVkvWFqNrRake 55
e ++k +yps++++ LA+++gL+ +q+ +WF N+R ++
Zjn_sc00014.1.g02690.1.am.mk 447 ETYYKWPYPSETQKVALAESTGLDLKQINNWFINQRKRH 485
568899******************************885 PP
| |||||||
| 2 | ELK | 36 | 1.5e-12 | 405 | 426 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELKh+Ll+KYsgyL+sLkqE+s
Zjn_sc00014.1.g02690.1.am.mk 405 ELKHHLLKKYSGYLSSLKQELS 426
9*******************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 8.3E-8 | 235 | 310 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 3.4E-15 | 236 | 309 | IPR005540 | KNOX1 |
| SMART | SM01256 | 2.3E-24 | 317 | 368 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 2.6E-20 | 323 | 366 | IPR005541 | KNOX2 |
| SMART | SM01188 | 570 | 326 | 346 | IPR005539 | ELK domain |
| SMART | SM01188 | 4.9E-7 | 405 | 426 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 11.331 | 405 | 425 | IPR005539 | ELK domain |
| Pfam | PF03789 | 1.8E-9 | 405 | 426 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 12.698 | 425 | 488 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.33E-18 | 426 | 501 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 2.1E-11 | 427 | 492 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 8.9E-26 | 430 | 491 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 1.10E-11 | 437 | 489 | No hit | No description |
| Pfam | PF05920 | 2.4E-14 | 445 | 484 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 463 | 486 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 708 aa Download sequence Send to blast |
MAPRPIPREH DDGRPSPHAC AVRLALPAHV LASQDYCQYH RNTSTSEGYT ATTPYHYRQR 60 QAPGSKGSSG DERIMAVPLG EASVYCALPR VFGYEIPYAY PCLLVTSPPA GRWLNGIIRL 120 LTDELGDSGR SARSLHQYIP GDPPAPMEEI SHNHFGGVGA SSHGQHQHNP WGSALSAVVG 180 PPPPLTLNTA ATGNSGASSG GNNPVLQLAN GGSLLDACVK AKEPSSSSPY AGDVEAIKAK 240 IISHPHYYSL LAAYLECQKA SPTLLLLLLH EKLLLHLMSN MEHAACNCKL VGAPPEVSAR 300 LTAMAQELEA RQRTSLLGLG SATEPELDQF MEAYHEMLVK FREELTRPLQ EAMEFMRRLE 360 LQLDSLSISS RSLRNILSSG SEEDQECSGG ETQLPEVDAH GVDQELKHHL LKKYSGYLSS 420 LKQELSKKKK KGKLPKEARQ KLLSWWETYY KWPYPSETQK VALAESTGLD LKQINNWFIN 480 QRKRHWKPSE EMHHMMMDGY HTSNAFYMDG HFFNEAGMYR DNSRIYTKGN DNEVELSRTY 540 LQCDGRWCPP KRRRHPPVLA PRLRRRNRPS EVRVRSITCL GWVCLCLALV PARTRARFGI 600 LGAMAPVHGV AGRCLPRADE AASGARDAAC PEFGHDAQAD SAAASVLQLQ VCKPLDGMPA 660 RTGITSESFL AFGFPICDAG YSLTCALANW SDRWERLLSS TGDDGVSP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Binds to RNA (PubMed:17965274). Possible transcription factor that regulates genes involved in development. Mutations in KN-1 alter leaf development. Foci of cells along the lateral vein do not differentiate properly but continue to divide, forming knots. May participate in the switch from indeterminate to determinate cell fates. Probably binds to the DNA sequence 5'-TGAC-3'. {ECO:0000269|PubMed:17965274}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00675 | ChIP-seq | Transfer from GRMZM2G017087 | Download |
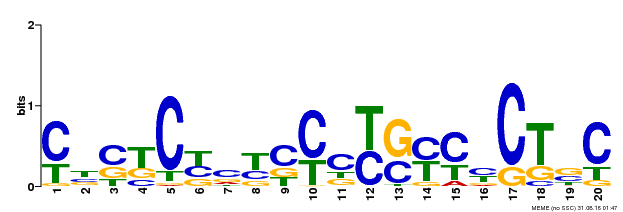
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | DQ317421 | 0.0 | DQ317421.1 Chasmanthium latifolium KNOTTED1 homeodomain protein (KN1) mRNA, partial cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004981913.1 | 0.0 | homeotic protein knotted-1 | ||||
| Swissprot | P24345 | 0.0 | KN1_MAIZE; Homeotic protein knotted-1 | ||||
| TrEMBL | A0A368SF91 | 0.0 | A0A368SF91_SETIT; Uncharacterized protein | ||||
| STRING | Pavir.J00864.1.p | 0.0 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP9886 | 31 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G08150.1 | 1e-81 | KNOTTED-like from Arabidopsis thaliana | ||||



