 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00014.1.g05830.1.am.mk | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | TALE | ||||||||
| Protein Properties | Length: 488aa MW: 54923.5 Da PI: 8.9952 | ||||||||
| Description | TALE family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 26.5 | 1.1e-08 | 413 | 447 | 21 | 55 |
HSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHH CS
Homeobox 21 knrypsaeereeLAkklgLterqVkvWFqNrRake 55
+ +yps++e+ LA+ +gL+++q+ +WF N+R ++
Zjn_sc00014.1.g05830.1.am.mk 413 RWPYPSETEKIALAEATGLDQKQINNWFINQRKRH 447
679*****************************885 PP
| |||||||
| 2 | ELK | 40.5 | 5.8e-14 | 367 | 388 | 1 | 22 |
ELK 1 ELKhqLlrKYsgyLgsLkqEFs 22
ELKh+Ll+KY+gyLg+L+qEFs
Zjn_sc00014.1.g05830.1.am.mk 367 ELKHHLLKKYGGYLGGLRQEFS 388
9********************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01255 | 2.0E-19 | 234 | 278 | IPR005540 | KNOX1 |
| Pfam | PF03790 | 1.2E-21 | 235 | 276 | IPR005540 | KNOX1 |
| SMART | SM01256 | 4.2E-26 | 284 | 335 | IPR005541 | KNOX2 |
| Pfam | PF03791 | 5.3E-22 | 290 | 334 | IPR005541 | KNOX2 |
| Pfam | PF03789 | 1.5E-10 | 367 | 388 | IPR005539 | ELK domain |
| SMART | SM01188 | 1.5E-7 | 367 | 388 | IPR005539 | ELK domain |
| PROSITE profile | PS51213 | 11.505 | 367 | 387 | IPR005539 | ELK domain |
| PROSITE profile | PS50071 | 13.151 | 387 | 450 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 1.33E-19 | 388 | 458 | IPR009057 | Homeodomain-like |
| SMART | SM00389 | 2.9E-12 | 389 | 454 | IPR001356 | Homeobox domain |
| Gene3D | G3DSA:1.10.10.60 | 1.7E-27 | 392 | 453 | IPR009057 | Homeodomain-like |
| CDD | cd00086 | 2.21E-11 | 399 | 450 | No hit | No description |
| Pfam | PF05920 | 3.9E-16 | 407 | 446 | IPR008422 | Homeobox KN domain |
| PROSITE pattern | PS00027 | 0 | 425 | 448 | IPR017970 | Homeobox, conserved site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 488 aa Download sequence Send to blast |
MHRRVRPSRQ AERSEQRASE LIKLLRRDSI VDLTRPQQPG IRRGSRGNGQ GDGGCMAWAR 60 CVGRWPRDLT RPGRQLDPPP AIRAALRPDS GAQCWSVLLA SFSASSRHAR MFLYRYLVWS 120 FPLRWSLSIL AENSLAVRQA AVILRLTNTE GAMERFAELG GGGSSSTFNA SFLQLPLLAA 180 ASAQAIPPKA QQDRSRLALQ QLPADMSSSV PTVRHNHQKG GEVVMGEISP ADAETVKAEI 240 MSHPQYPALL TAYLDCQKVG APPDVSDRLS AVAAKVDTLQ WKDQRRPQRA NPELDQFMEA 300 YCNMLVKYQE ELARPILEAA EFFNSVEKQL DSITDSSTRD GAGSSDVDDD QDASCPEELD 360 PCAEDKELKH HLLKKYGGYL GGLRQEFSKR KKKGKLPKEA RQKLLQWWEL HYRWPYPSET 420 EKIALAEATG LDQKQINNWF INQRKRHWKP TSEDMPPFAM VMEGCFHAPP ALYMDRPFMA 480 DGMYRLGP |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that may be involved in shoot formation during embryogenesis. {ECO:0000269|PubMed:10080693}. | |||||
| UniProt | Probable transcription factor that may be involved in shoot formation during embryogenesis. {ECO:0000269|PubMed:10080693, ECO:0000269|PubMed:9869405}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00671 | PBM | Transfer from GRMZM2G135447 | Download |
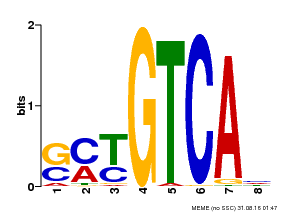
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025795146.1 | 1e-155 | homeobox protein knotted-1-like 12 isoform X3 | ||||
| Swissprot | O65034 | 1e-105 | KNOSC_ORYSI; Homeobox protein knotted-1-like 12 | ||||
| Swissprot | O80416 | 1e-105 | KNOSC_ORYSJ; Homeobox protein knotted-1-like 12 | ||||
| TrEMBL | A0A0A9GAR1 | 1e-173 | A0A0A9GAR1_ARUDO; Uncharacterized protein | ||||
| STRING | Pavir.Ia00428.1.p | 1e-147 | (Panicum virgatum) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4061 | 38 | 73 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G08150.1 | 1e-84 | KNOTTED-like from Arabidopsis thaliana | ||||



