 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00022.1.g06600.1.am.mk | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 475aa MW: 50955.6 Da PI: 8.406 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 55.3 | 1.7e-17 | 317 | 366 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GVr + +g+Wv+eIr+p++ + r++lg+f tae+Aa+a++ a+++++g
Zjn_sc00022.1.g06600.1.am.mk 317 TYRGVRMRA-WGKWVSEIREPRK---KSRIWLGTFATAEMAARAHDVAALAIKG 366
69****998.**********843...6***********************9987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF54171 | 7.19E-21 | 317 | 375 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 3.3E-37 | 317 | 380 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 9.9E-32 | 317 | 375 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 22.958 | 317 | 374 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 9.0E-12 | 317 | 366 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 1.58E-28 | 318 | 376 | No hit | No description |
| PRINTS | PR00367 | 4.4E-10 | 318 | 329 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 4.4E-10 | 340 | 356 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 475 aa Download sequence Send to blast |
MEDKVRWGGE GRKKVQFGAA VECTTESGVP VVARPAKIAR DRPSLADRQH MRVRSTTILT 60 CDREIESDGV DLRYISPEAW RLRRTTTCTA NEQSVERKLN ELHATIDCYD PNKSNPTERA 120 SELAGSSAYS KRRYETRGRV GRAESEARAK GHGRHHGGVE TNTHGAGASS WHAWPFGPSR 180 TTRLRRRSSV RSNPAGLPAA LHWGRGPCSS IAPVDLRETV DGDPCRKSPT APLVSSAHCH 240 TPPSSALLLT NSTTNTNTSS AAMAMDDPSS GSETTTSSSA EAPASPTATA TSASSSDAAA 300 EGVGKKKRRR SDGHHPTYRG VRMRAWGKWV SEIREPRKKS RIWLGTFATA EMAARAHDVA 360 ALAIKGRAAH LNFPELASEL PRPATAAPKD VQAAAALAAA ADFLVPAANA GAKDAEEPEA 420 SPASASPPQD NAEDALFDLP DLLFDLRYGP PSCAASWEDD GAGVFRLEEP LLWEY |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 2e-16 | 297 | 374 | 2 | 72 | Ethylene-responsive transcription factor ERF096 |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 304 | 309 | KKKRRR |
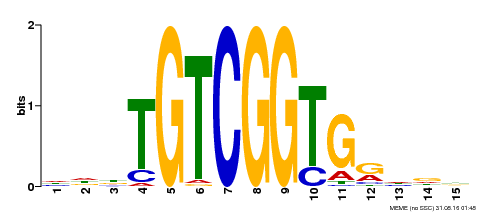
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00312 | DAP | Transfer from AT2G44940 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KJ728262 | 1e-110 | KJ728262.1 Zea mays clone pUT6539 AP2-EREBP transcription factor (EREB176) gene, partial cds. | |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP159 | 35 | 354 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G44940.1 | 2e-38 | ERF family protein | ||||



