 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00039.1.g01260.1.am.mk | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | HSF | ||||||||
| Protein Properties | Length: 850aa MW: 91457.5 Da PI: 10.0549 | ||||||||
| Description | HSF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HSF_DNA-bind | 110.8 | 9.5e-35 | 507 | 600 | 2 | 103 |
HHHHHHHHHCTGGGTTTSEES.SSSSEEEES-HHHHHHHTHHHHSTT--HHHHHHHHHHTTEEE---SSBTTTTXTTSEEE CS
HSF_DNA-bind 2 Flkklyeiledeelkelisws.engnsfvvldeeefakkvLpkyFkhsnfaSFvRQLnmYgFkkvkdeekkskskekiweF 81
Fl+k+y++++d+++++++sw ++ ++fvv+ + efa+++Lp+yFkh+nf+SFvRQLn+YgF+kv e+ weF
Zjn_sc00039.1.g01260.1.am.mk 507 FLTKTYQLVDDPATDHIVSWGdDRVSTFVVWRPPEFARDILPNYFKHNNFSSFVRQLNTYGFRKVVPER---------WEF 578
9********************6667*****************************************999.........*** PP
EESXXXXXXXXXXXXXXXXXXX CS
HSF_DNA-bind 82 khksFkkgkkellekikrkkse 103
+++ F+kg+k+ll++i+r+k++
Zjn_sc00039.1.g01260.1.am.mk 579 ANEFFRKGDKQLLCEIHRRKTS 600
*******************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 9.5E-36 | 497 | 592 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM00415 | 7.1E-58 | 503 | 597 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| SuperFamily | SSF46785 | 1.36E-30 | 504 | 597 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| PRINTS | PR00056 | 1.4E-18 | 507 | 530 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Pfam | PF00447 | 5.7E-30 | 507 | 597 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 1.4E-18 | 546 | 558 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PROSITE pattern | PS00434 | 0 | 547 | 571 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| PRINTS | PR00056 | 1.4E-18 | 559 | 571 | IPR000232 | Heat shock factor (HSF)-type, DNA-binding |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0008356 | Biological Process | asymmetric cell division | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 850 aa Download sequence Send to blast |
MQPPCGAVDH PPSPVTGNSP CRSSPGPWAV YSEGGGRQQR PQAQHSKRTR RGAFEEGRGP 60 CTRSDRQMQS HYSSSVLAKP ASDAMHLRVS CRPDGRDASG RVPTKERRRR FRWWDSAGQP 120 VVIVLSLPIT GAAGQREVPD VAMLIIPSQA LLMHTVSPLS VRCCMQRHRP GCKRAGRGRR 180 YYQYVVNRRH SPPDRFRNPA AKGTPLQRCR IPHEGGPGGR SQAITPGPAA SALTVTATPD 240 RPATIRDRSC KFVIGTTGRR ASAAAQCPVI SGQRASEEGT ARHVEAESDD ARCPGAESGI 300 DGPAAAAPRA VDRAPGPSSP QFRTGQAGAC WPKPYPLQSR YEYKGQFPGP AAPATARDVH 360 RSGHMERCSD PRSAATQYRD SKNHIPILQY STIAACPPGT FAVLHGDTGG ARAGKMSDLI 420 DPCTEADRQS DTYSLPPPPP PARSPGLGHT PHLPSGVTSS DSLRHRPAPV ENGGKDRERM 480 ERVERGGSWE CDAAARSAAQ KAVPAPFLTK TYQLVDDPAT DHIVSWGDDR VSTFVVWRPP 540 EFARDILPNY FKHNNFSSFV RQLNTYGFRK VVPERWEFAN EFFRKGDKQL LCEIHRRKTS 600 SASTASPSPP PFFAPPHFPL FHPGVQHHHH QFVGDDGVVA AHGVGVGFPH PHWREQHAAT 660 RLLALGAPAT SPSSATDGGN CRAATAAVLM EENERLRRSN TALLQELAHM RKLYNDIIYF 720 VQNHVRPVAP SPAATAFLQG LGLQARNKPV GTALNNSGGS TTSSSSLTIA EEPSPPPQQL 780 LDKESGSSAP TKLFGVQLTA PGGAGTKRPP SPDEDQPPSS PATKPRLVLE SADLSLSTAP 840 SAASSPASTS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2ldu_A | 4e-21 | 498 | 600 | 12 | 123 | Heat shock factor protein 1 |
| 5hdg_A | 3e-21 | 503 | 599 | 7 | 112 | Heat shock factor protein 1 |
| 5hdn_A | 3e-21 | 503 | 599 | 7 | 112 | Heat shock factor protein 1 |
| 5hdn_B | 3e-21 | 503 | 599 | 7 | 112 | Heat shock factor protein 1 |
| 5hdn_C | 3e-21 | 503 | 599 | 7 | 112 | Heat shock factor protein 1 |
| 5hdn_D | 3e-21 | 503 | 599 | 7 | 112 | Heat shock factor protein 1 |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 105 | 110 | ERRRRF |
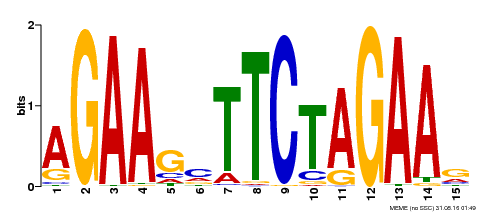
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00187 | DAP | Transfer from AT1G46264 | Download |
| |||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP121 | 37 | 394 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G46264.1 | 1e-58 | heat shock transcription factor B4 | ||||



