 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00040.1.g03480.1.sm.mkhc | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | WOX | ||||||||
| Protein Properties | Length: 274aa MW: 30878.3 Da PI: 6.1825 | ||||||||
| Description | WOX family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 66.3 | 4e-21 | 91 | 151 | 2 | 57 |
T--SS--HHHHHHHHHHHHH.SSS--HHHHHHHHHHC....TS-HHHHHHHHHHHHHHHHC CS
Homeobox 2 rkRttftkeqleeLeelFek.nrypsaeereeLAkkl....gLterqVkvWFqNrRakekk 57
r+R+++t+ ql++Le+ F + n +ps+++++e+++ l +++e++V++WFqNrRa+ k+
Zjn_sc00040.1.g03480.1.sm.mkhc 91 RQRWQPTPMQLQILENIFDQgNGTPSKQKIKEITADLshhgQISETNVYNWFQNRRARSKR 151
89*****************99**************************************97 PP
| |||||||
| 2 | Wus_type_Homeobox | 100.4 | 1.3e-32 | 90 | 153 | 2 | 65 |
Wus_type_Homeobox 2 artRWtPtpeQikiLeelyksGlrtPnkeeiqritaeLeeyGkiedkNVfyWFQNrkaRerqkq 65
+r+RW Ptp Q++iLe+++++G tP+k++i++ita L+++G+i+++NV++WFQNr+aR+++kq
Zjn_sc00040.1.g03480.1.sm.mkhc 90 TRQRWQPTPMQLQILENIFDQGNGTPSKQKIKEITADLSHHGQISETNVYNWFQNRRARSKRKQ 153
789***********************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 5.0E-17 | 86 | 157 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 14.803 | 87 | 152 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 5.6E-14 | 89 | 156 | IPR001356 | Homeobox domain |
| SuperFamily | SSF46689 | 3.89E-18 | 90 | 155 | IPR009057 | Homeodomain-like |
| Pfam | PF00046 | 6.4E-19 | 91 | 151 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 3.79E-13 | 91 | 153 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 274 aa Download sequence Send to blast |
MEWGDRATAA AAATEEAEER GGEPLGYVKV MTDEQMEVLR KQISIYATIC GQLVEMHRAL 60 AEHQDSIAGM GFSNLYCDPL IVPGGHKITT RQRWQPTPMQ LQILENIFDQ GNGTPSKQKI 120 KEITADLSHH GQISETNVYN WFQNRRARSK RKQAAALPNN VESEAEADEE SLTDKKPNSD 180 MQLQQSKALN VHNADRFSEM HHIQRDQMSA MMYGSNDNNL RSSGSSGQMS FYENIMSNPR 240 IDQFSGKVEN SRSFSHLQHG EGFDIGWSRE KKGS |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 144 | 152 | RRARSKRKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor which may be involved in developmental processes. {ECO:0000250}. | |||||
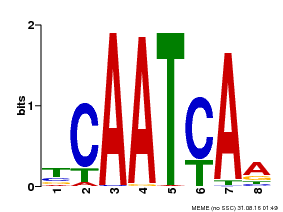
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00669 | PBM | Transfer from GRMZM2G038252 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_025815130.1 | 1e-154 | WUSCHEL-related homeobox 8-like | ||||
| Swissprot | Q5QMM3 | 1e-120 | WOX8_ORYSJ; WUSCHEL-related homeobox 8 | ||||
| TrEMBL | A0A2S3HQI6 | 1e-152 | A0A2S3HQI6_9POAL; Uncharacterized protein | ||||
| STRING | Sb03g038080.1 | 1e-153 | (Sorghum bicolor) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP3898 | 37 | 76 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G35550.1 | 1e-54 | WUSCHEL related homeobox 13 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



