 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zjn_sc00106.1.g01220.1.am.mk | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 367aa MW: 40272 Da PI: 10.0994 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 62.4 | 1e-19 | 235 | 284 | 2 | 55 |
AP2 2 gykGVrwdkkrgrWvAeIrdpsengkr.krfslgkfgtaeeAakaaiaarkkleg 55
+++GVr+++ +g+W+AeIrdp+ r r +lg+f+taeeAaka+++a+ +++g
Zjn_sc00106.1.g01220.1.am.mk 235 RFRGVRQRP-WGKWAAEIRDPR----RaVRKWLGTFDTAEEAAKAYDRAAVEFRG 284
69*******.**********82....48***********************9998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51032 | 24.658 | 235 | 292 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 5.82E-22 | 235 | 293 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 1.5E-30 | 235 | 292 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 1.8E-36 | 235 | 298 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 8.6E-13 | 235 | 284 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 1.6E-10 | 236 | 247 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 7.19E-27 | 238 | 292 | No hit | No description |
| PRINTS | PR00367 | 1.6E-10 | 258 | 274 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0001944 | Biological Process | vasculature development | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0051301 | Biological Process | cell division | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 367 aa Download sequence Send to blast |
MSSVNSRDAT PLAFTLPRIQ LEAKTGIRKK TEDGRRARFS RLSRALSTQR PRYSPRPLRR 60 DPRIKAPAPP ASSRSIQASA TTRNADNRMT NRTFLPSPSM AASAANQDYV IRFDGQFDDP 120 SPSSSSGHEP PPPPFAWRPI TPEREHAVIV AALLHVVSGY TTPPPEVFPA AQVEASTSCR 180 VCGMERCLGC EFFAADTFSA GADTAENAVV TAAATTTAGA GGTGTQRRRR KKKNRFRGVR 240 QRPWGKWAAE IRDPRRAVRK WLGTFDTAEE AAKAYDRAAV EFRGPRAKLN FPFPEHQAAL 300 TTAHDAAAAK SSDTQSPCSA DVEDRSGPQD WAPRGGEEAG ELWDDLQDLM RLDEGEIWFP 360 PTSTPWN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2gcc_A | 3e-20 | 232 | 293 | 2 | 64 | ATERF1 |
| 3gcc_A | 3e-20 | 232 | 293 | 2 | 64 | ATERF1 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 226 | 232 | RRRRKKK |
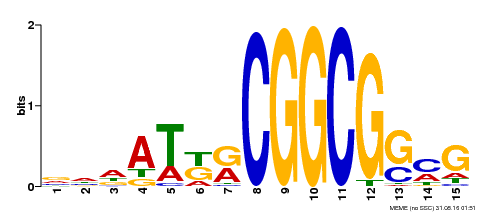
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00469 | DAP | Transfer from AT4G34410 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU964453 | 4e-67 | EU964453.1 Zea mays clone 278992 AP2 domain containing protein mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004973673.1 | 1e-110 | ethylene-responsive transcription factor ERF109 | ||||
| TrEMBL | K3YJ18 | 1e-108 | K3YJ18_SETIT; Uncharacterized protein | ||||
| STRING | Si014237m | 1e-109 | (Setaria italica) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP1242 | 36 | 124 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G34410.1 | 7e-18 | redox responsive transcription factor 1 | ||||



