 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Zmw_sc00918.1.g00040.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Chloridoideae; Zoysieae; Zoysiinae; Zoysia
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 704aa MW: 75837.8 Da PI: 6.5652 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 14.8 | 5.4e-05 | 538 | 559 | 4 | 25 |
HHHHHHHHHHHHHHHHHHHHHC CS
HLH 4 ahnerErrRRdriNsafeeLre 25
+h e+Er+RR+++N++f Lr
Zmw_sc00918.1.g00040.1.am.mk 538 NHVEAERQRREKLNQRFYALRA 559
799**************99996 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 9.0E-52 | 93 | 273 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 9.343 | 534 | 591 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 3.27E-10 | 537 | 596 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 6.34E-8 | 537 | 578 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 9.3E-6 | 538 | 596 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 5.7E-4 | 540 | 580 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009269 | Biological Process | response to desiccation | ||||
| GO:0009611 | Biological Process | response to wounding | ||||
| GO:0009737 | Biological Process | response to abscisic acid | ||||
| GO:0009867 | Biological Process | jasmonic acid mediated signaling pathway | ||||
| GO:0009963 | Biological Process | positive regulation of flavonoid biosynthetic process | ||||
| GO:0010200 | Biological Process | response to chitin | ||||
| GO:0043619 | Biological Process | regulation of transcription from RNA polymerase II promoter in response to oxidative stress | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0051090 | Biological Process | regulation of sequence-specific DNA binding transcription factor activity | ||||
| GO:2000068 | Biological Process | regulation of defense response to insect | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 704 aa Download sequence Send to blast |
MPPLIKRASV LFWDISLAFP SAPSVSTISP LAPVPRPSMN LWTDDNASMM EAFMASADLT 60 WGTVGGDGSA GATPPPPFHQ MPAAAPAFNQ DTLQQRLQAI IEGSRETWTY AIYWQSSVDA 120 GSGASLLGWG DGYYKGCDED KRRQRPLTPA DQVEQEHRKR VLRELNSLIS GGAPAPDEAV 180 EEEVTDTEWF FLVSMTQSFS HGSGLPAQVF FSGQPNWISS GLSAAPCERA RQAYTFGLRT 240 MVCVPVGTGV LELGSTDVIF HTAESMGKIR SLFGGGAGPG SWQPQQQPAA GAEHAETDLW 300 LADTPATDMK DTMSHPAAEI SVSKPPQIHF ENVTTTTSTL TENPSPSVQA PSAPQPAVPV 360 PPQRQQQQQQ SHVQQGPFRR ELNFSEFASN PSVVSGPPFF KPESGEILSF GADSTSRRNP 420 SPAPPTTTAP GSLFSQNTAT ITAAATNDAK NNNKRSMEAT SRASNTNLHP APTANEGMLS 480 FSSAPTTRPS TGTGAPAKSE SDHSDLDARV VAPPPPEAEK RPRKRGRKPA NGREEPLNHV 540 EAERQRREKL NQRFYALRAV ASLLGDAISY INELRGKLTA LESDKETMQF QIEALKKERD 600 ARPAPQAGGF GHDAGPRCHA CHKRNHPSAR LMTALRELDL DVYHASVSVV AVKMASRIYS 660 QDQLNAALYS RLAEPGTVMG RNDTVTLTHL LLVVVVITVN SGMA |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rqw_A | 4e-61 | 86 | 273 | 2 | 192 | Transcription factor MYC3 |
| 4rqw_B | 4e-61 | 86 | 273 | 2 | 192 | Transcription factor MYC3 |
| 4rs9_A | 4e-61 | 86 | 273 | 2 | 192 | Transcription factor MYC3 |
| 4yz6_A | 4e-61 | 86 | 273 | 2 | 192 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 519 | 527 | KRPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator involved in jasmonate (JA) signaling pathway during spikelet development. Binds to the G2 region G-box (5'-CACGTG-3') of the MADS1 promoter and thus directly regulates the expression of MADS1. Its function in MADS1 activation is abolished by TIFY3/JAZ1 which directly target MYC2 during spikelet development. {ECO:0000269|PubMed:24647160}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00084 | PBM | Transfer from AT1G32640 | Download |
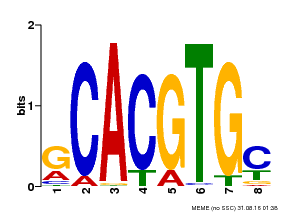
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_012698381.1 | 0.0 | LOW QUALITY PROTEIN: transcription factor MYC2-like | ||||
| Swissprot | Q336P5 | 0.0 | MYC2_ORYSJ; Transcription factor MYC2 | ||||
| TrEMBL | A0A1E5W733 | 0.0 | A0A1E5W733_9POAL; Transcription factor MYC2 | ||||
| STRING | GRMZM2G001930_P01 | 0.0 | (Zea mays) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Monocots | OGMP4452 | 31 | 57 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



